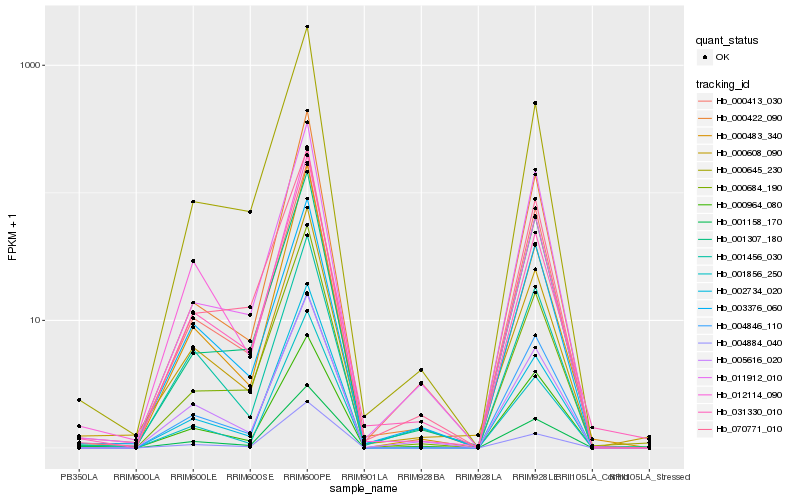
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000964_080 |
0.0728471481 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 3 |
Hb_000608_090 |
0.0811374619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004846_110 |
0.0853763727 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 5 |
Hb_000684_190 |
0.0861648571 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000645_230 |
0.0893111719 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 7 |
Hb_000422_090 |
0.0896347866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005616_020 |
0.0909458922 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_004884_040 |
0.0942093248 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 10 |
Hb_031330_010 |
0.0955551573 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 11 |
Hb_003376_060 |
0.0961416333 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002734_020 |
0.0968516573 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 13 |
Hb_001856_250 |
0.0992262441 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 14 |
Hb_011912_010 |
0.0992989784 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_012114_090 |
0.0993373632 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 16 |
Hb_000483_340 |
0.0994008979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_070771_010 |
0.1004804775 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 18 |
Hb_000413_030 |
0.101838645 |
- |
- |
PREDICTED: NAD(P)H dehydrogenase (quinone) FQR1-like [Jatropha curcas] |
| 19 |
Hb_001456_030 |
0.1021982539 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 20 |
Hb_001307_180 |
0.1048574292 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |