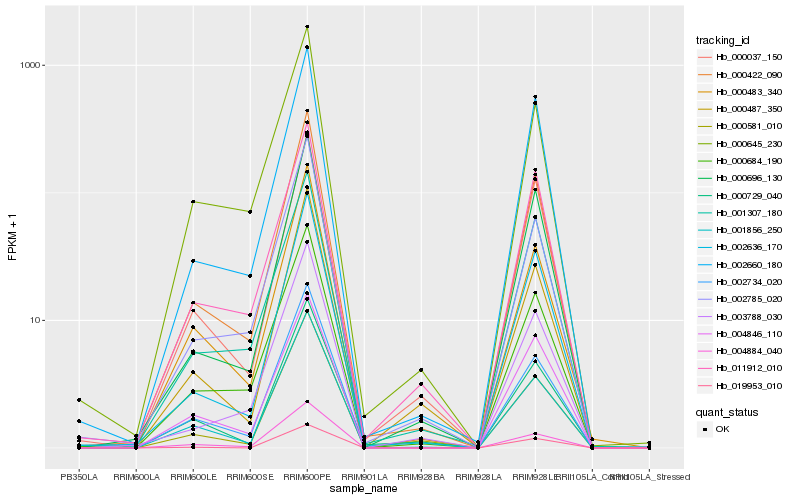
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000422_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002636_170 |
0.0412349276 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 3 |
Hb_000696_130 |
0.0480762551 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_002660_180 |
0.0562338403 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000684_190 |
0.0572647502 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 6 |
Hb_002734_020 |
0.0602161512 |
- |
- |
PREDICTED: uncharacterized protein LOC105631540 [Jatropha curcas] |
| 7 |
Hb_001856_250 |
0.0621376131 |
- |
- |
PREDICTED: phenolic glucoside malonyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_000037_150 |
0.068168119 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 9 |
Hb_002785_020 |
0.0694709876 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 10 |
Hb_000483_340 |
0.0706722853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000645_230 |
0.072898314 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 12 |
Hb_011912_010 |
0.0729125928 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 13 |
Hb_003788_030 |
0.0732323129 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_004846_110 |
0.0741068747 |
- |
- |
hypothetical protein JCGZ_04591 [Jatropha curcas] |
| 15 |
Hb_004884_040 |
0.0746223162 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 16 |
Hb_000729_040 |
0.0771095171 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 17 |
Hb_001307_180 |
0.0779507007 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 4 [UDP-forming] isoform X1 [Jatropha curcas] |
| 18 |
Hb_000581_010 |
0.0788114465 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like [Jatropha curcas] |
| 19 |
Hb_000487_350 |
0.0789017694 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_019953_010 |
0.0794940665 |
- |
- |
hypothetical protein CICLE_v10019877mg [Citrus clementina] |