| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
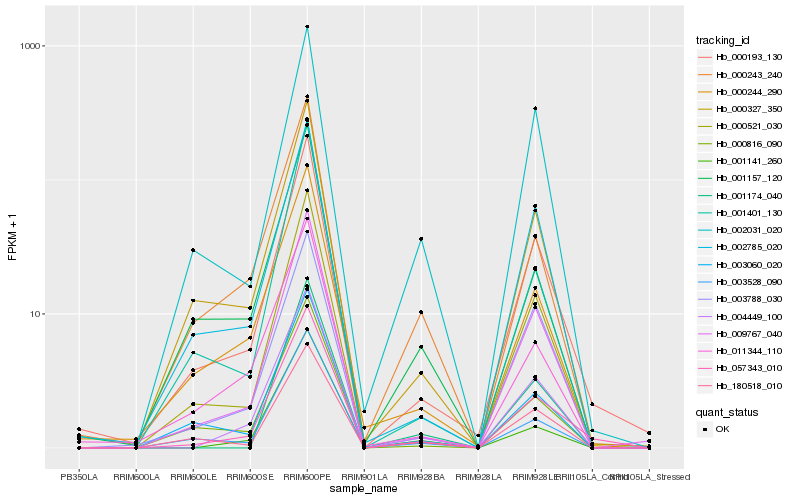
Hb_057343_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 2 |
Hb_000193_130 |
0.0782585304 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_004449_100 |
0.1011853608 |
- |
- |
- |
| 4 |
Hb_009767_040 |
0.1061898596 |
- |
- |
PREDICTED: uncharacterized protein LOC105127022 [Populus euphratica] |
| 5 |
Hb_000521_030 |
0.1085288277 |
- |
- |
PREDICTED: uncharacterized protein LOC105650350 [Jatropha curcas] |
| 6 |
Hb_000327_350 |
0.1117100852 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 7 |
Hb_003528_090 |
0.1182201507 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 8 |
Hb_000244_290 |
0.1200393524 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_000243_240 |
0.1239139659 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 10 |
Hb_000816_090 |
0.1273648004 |
transcription factor |
TF Family: TRAF |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_003060_020 |
0.1282798721 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 12 |
Hb_001401_130 |
0.1312696803 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 13 |
Hb_001141_260 |
0.131829334 |
- |
- |
Uncharacterized protein TCM_020647 [Theobroma cacao] |
| 14 |
Hb_002785_020 |
0.1323400979 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 15 |
Hb_011344_110 |
0.1323702705 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 16 |
Hb_180518_010 |
0.1357548417 |
- |
- |
PREDICTED: receptor-like protein 2 [Jatropha curcas] |
| 17 |
Hb_001174_040 |
0.1369004928 |
- |
- |
- |
| 18 |
Hb_002031_020 |
0.1380138133 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 19 |
Hb_003788_030 |
0.1381884785 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_001157_120 |
0.1401998494 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |