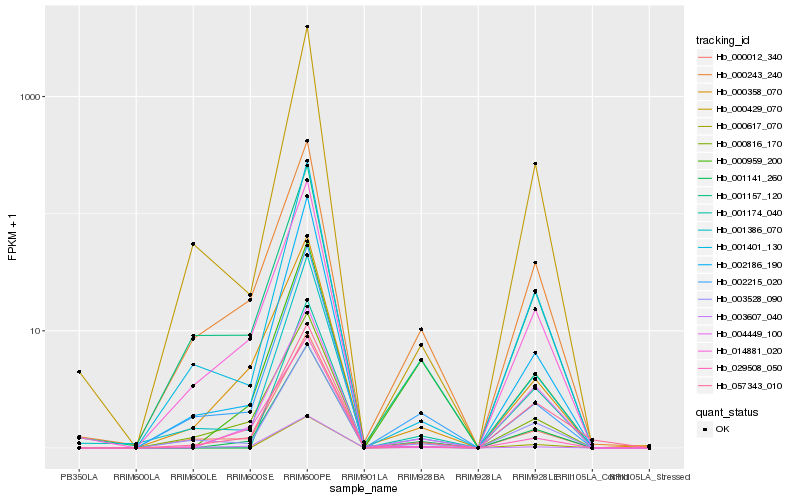
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_260 |
0.0 |
- |
- |
Uncharacterized protein TCM_020647 [Theobroma cacao] |
| 2 |
Hb_000617_070 |
0.101619027 |
- |
- |
hypothetical protein POPTR_0005s02420g [Populus trichocarpa] |
| 3 |
Hb_000243_240 |
0.1053385327 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 4 |
Hb_004449_100 |
0.1090613715 |
- |
- |
- |
| 5 |
Hb_000959_200 |
0.1114167038 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 6 |
Hb_002186_190 |
0.1136166166 |
- |
- |
- |
| 7 |
Hb_002215_020 |
0.1143101862 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 8 |
Hb_000816_170 |
0.1158442409 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 15 [Jatropha curcas] |
| 9 |
Hb_000358_070 |
0.1162708013 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 10 |
Hb_001401_130 |
0.1174239859 |
- |
- |
Calcium-binding EF-hand family protein, putative [Theobroma cacao] |
| 11 |
Hb_003528_090 |
0.1176873851 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |
| 12 |
Hb_000012_340 |
0.1222522127 |
- |
- |
PREDICTED: inorganic pyrophosphatase 3-like [Jatropha curcas] |
| 13 |
Hb_001157_120 |
0.1235653552 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 14 |
Hb_001174_040 |
0.1260459754 |
- |
- |
- |
| 15 |
Hb_000429_070 |
0.1275344769 |
- |
- |
hypothetical protein POPTR_0006s05480g [Populus trichocarpa] |
| 16 |
Hb_014881_020 |
0.1285450457 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 17 |
Hb_003607_040 |
0.1287171844 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 18 |
Hb_001386_070 |
0.1299986927 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1-like [Jatropha curcas] |
| 19 |
Hb_057343_010 |
0.131829334 |
- |
- |
hypothetical protein JCGZ_23164 [Jatropha curcas] |
| 20 |
Hb_029508_050 |
0.1327356451 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like isoform X2 [Musa acuminata subsp. malaccensis] |