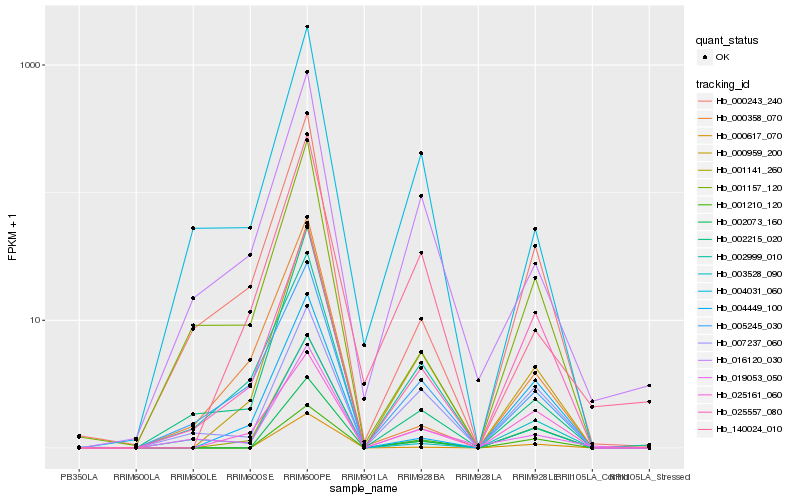
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 2 |
Hb_001141_260 |
0.1114167038 |
- |
- |
Uncharacterized protein TCM_020647 [Theobroma cacao] |
| 3 |
Hb_140024_010 |
0.1158308645 |
- |
- |
PREDICTED: uncharacterized protein C167.05 [Vitis vinifera] |
| 4 |
Hb_016120_030 |
0.1208675699 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |
| 5 |
Hb_004031_060 |
0.1301071341 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |
| 6 |
Hb_005245_030 |
0.1307804522 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 7 |
Hb_019053_050 |
0.1435822098 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_000243_240 |
0.15094076 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 9 |
Hb_002215_020 |
0.1535465024 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 10 |
Hb_002999_010 |
0.1569355881 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 11 |
Hb_001210_120 |
0.1573634902 |
- |
- |
hypothetical protein JCGZ_06226 [Jatropha curcas] |
| 12 |
Hb_025557_080 |
0.1597684103 |
transcription factor |
TF Family: zf-HD |
- |
| 13 |
Hb_000617_070 |
0.1633654706 |
- |
- |
hypothetical protein POPTR_0005s02420g [Populus trichocarpa] |
| 14 |
Hb_025161_060 |
0.1668224558 |
- |
- |
hypothetical protein JCGZ_23770 [Jatropha curcas] |
| 15 |
Hb_001157_120 |
0.1698608238 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 16 |
Hb_002073_160 |
0.1700501092 |
- |
- |
hypothetical protein JCGZ_21740 [Jatropha curcas] |
| 17 |
Hb_007237_060 |
0.171094629 |
- |
- |
- |
| 18 |
Hb_004449_100 |
0.1773747958 |
- |
- |
- |
| 19 |
Hb_000358_070 |
0.1783004213 |
- |
- |
hypothetical protein POPTR_0003s01410g [Populus trichocarpa] |
| 20 |
Hb_003528_090 |
0.1795128885 |
- |
- |
Uncharacterized protein TCM_037169 [Theobroma cacao] |