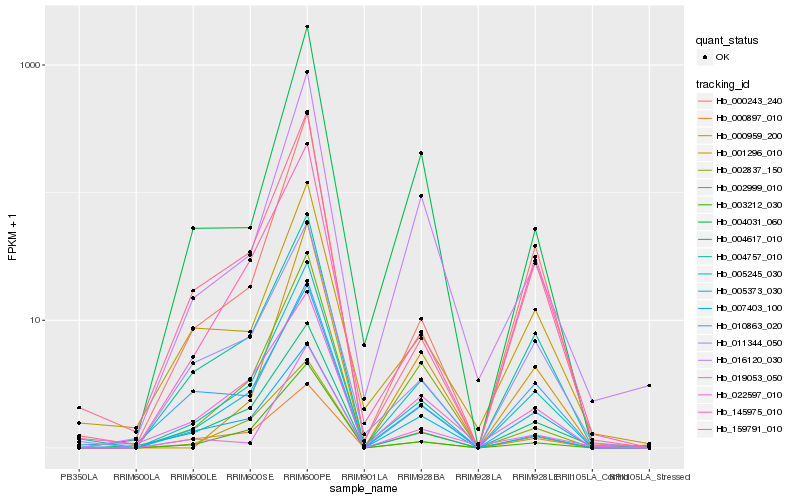
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005245_030 |
0.0 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At3g07570 [Jatropha curcas] |
| 2 |
Hb_016120_030 |
0.1082598099 |
- |
- |
hypothetical protein EUGRSUZ_D00080 [Eucalyptus grandis] |
| 3 |
Hb_005373_030 |
0.1126532236 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002999_010 |
0.1152543809 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 5 |
Hb_004031_060 |
0.1170493393 |
- |
- |
hypothetical protein B456_007G3219002, partial [Gossypium raimondii] |
| 6 |
Hb_022597_010 |
0.1229018292 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 13 [Vitis vinifera] |
| 7 |
Hb_004617_010 |
0.1271904614 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X3 [Citrus sinensis] |
| 8 |
Hb_000243_240 |
0.1278567196 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 9 |
Hb_011344_050 |
0.1280676366 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 10 |
Hb_003212_030 |
0.1293705897 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 11 |
Hb_000959_200 |
0.1307804522 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 12 |
Hb_145975_010 |
0.1325132867 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_001296_010 |
0.1350315586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000897_010 |
0.1360905531 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 15 |
Hb_004757_010 |
0.1382001214 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 16 |
Hb_007403_100 |
0.1383220902 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 17 |
Hb_002837_150 |
0.1388845947 |
- |
- |
hypothetical protein EUGRSUZ_L00324 [Eucalyptus grandis] |
| 18 |
Hb_159791_010 |
0.1391415468 |
- |
- |
PREDICTED: cytochrome P450 84A1 [Jatropha curcas] |
| 19 |
Hb_010863_020 |
0.1418083309 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g03010-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_019053_050 |
0.1421390928 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |