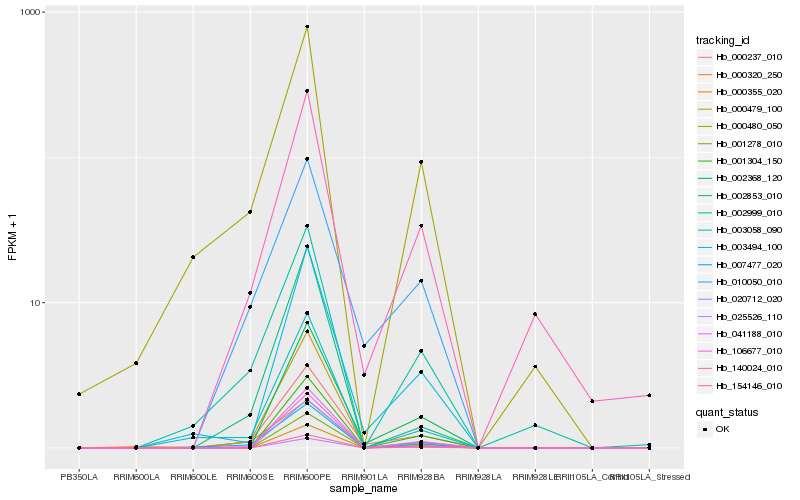
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_010 |
0.0 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010050_010 |
0.1019932567 |
- |
- |
- |
| 3 |
Hb_007477_020 |
0.1241550464 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA5 [Jatropha curcas] |
| 4 |
Hb_003494_100 |
0.1315786017 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 1 [Theobroma cacao] |
| 5 |
Hb_140024_010 |
0.1362263835 |
- |
- |
PREDICTED: uncharacterized protein C167.05 [Vitis vinifera] |
| 6 |
Hb_002368_120 |
0.1383844271 |
- |
- |
hypothetical protein VITISV_033181 [Vitis vinifera] |
| 7 |
Hb_106677_010 |
0.1506905203 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002853_010 |
0.1541519344 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000479_100 |
0.1555297757 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic [Gossypium arboreum] |
| 10 |
Hb_003058_090 |
0.1569266312 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 11 |
Hb_002999_010 |
0.1592677571 |
- |
- |
PREDICTED: extracellular ribonuclease LE-like [Vitis vinifera] |
| 12 |
Hb_000480_050 |
0.1606766329 |
- |
- |
hypothetical protein PHAVU_004G154200g [Phaseolus vulgaris] |
| 13 |
Hb_020712_020 |
0.1641472153 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Elaeis guineensis] |
| 14 |
Hb_000355_020 |
0.1641909048 |
- |
- |
- |
| 15 |
Hb_001278_010 |
0.1642837836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_025526_110 |
0.1645783493 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 17 |
Hb_041188_010 |
0.1648608262 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Nicotiana tomentosiformis] |
| 18 |
Hb_154146_010 |
0.1664554912 |
- |
- |
hypothetical protein JCGZ_00346 [Jatropha curcas] |
| 19 |
Hb_001304_150 |
0.1669474599 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_000320_250 |
0.1693036854 |
- |
- |
hypothetical protein POPTR_0021s00440g [Populus trichocarpa] |