| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
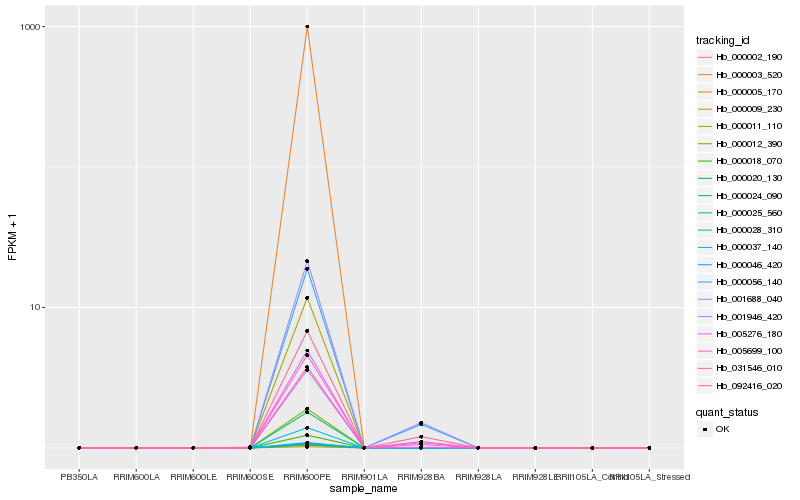
Hb_005276_180 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s23040g [Populus trichocarpa] |
| 2 |
Hb_001688_040 |
0.0259816576 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000056_140 |
0.0293438276 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE6 [Vitis vinifera] |
| 4 |
Hb_001946_420 |
0.0454423481 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_092416_020 |
0.0489609189 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_031546_010 |
0.0600943494 |
- |
- |
hypothetical protein JCGZ_21533 [Jatropha curcas] |
| 7 |
Hb_005699_100 |
0.0731350431 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 8 |
Hb_000002_190 |
0.0895913871 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 9 |
Hb_000003_520 |
0.0895913871 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 10 |
Hb_000005_170 |
0.0895913871 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR [Jatropha curcas] |
| 11 |
Hb_000009_230 |
0.0895913871 |
- |
- |
hypothetical protein POPTR_0007s12290g [Populus trichocarpa] |
| 12 |
Hb_000011_110 |
0.0895913871 |
- |
- |
PREDICTED: uncharacterized protein LOC105631299 [Jatropha curcas] |
| 13 |
Hb_000012_390 |
0.0895913871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000018_070 |
0.0895913871 |
- |
- |
PREDICTED: amino acid permease 8-like [Jatropha curcas] |
| 15 |
Hb_000020_130 |
0.0895913871 |
- |
- |
conidial pigment biosynthesis oxidase Arb2/brown2 [Aspergillus clavatus NRRL 1] |
| 16 |
Hb_000024_090 |
0.0895913871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000025_560 |
0.0895913871 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_000028_310 |
0.0895913871 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 19 |
Hb_000037_140 |
0.0895913871 |
- |
- |
PREDICTED: uncharacterized protein LOC104586896 [Nelumbo nucifera] |
| 20 |
Hb_000046_420 |
0.0895913871 |
- |
- |
hypothetical protein JCGZ_02014 [Jatropha curcas] |