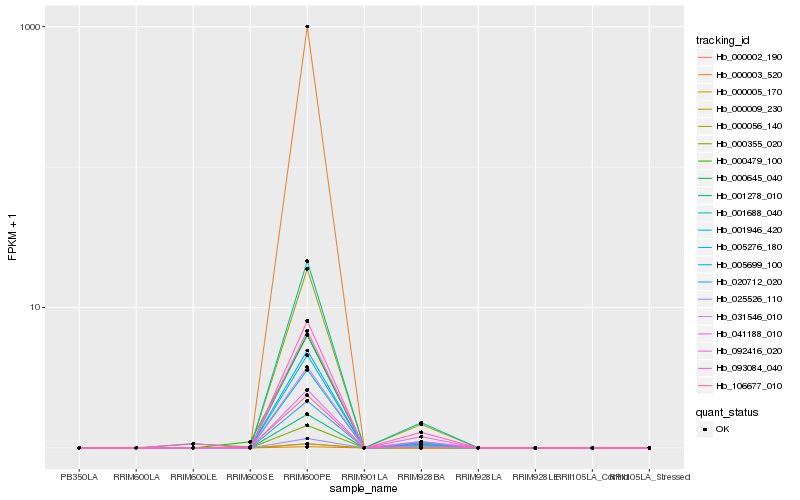
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001688_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000056_140 |
0.0033821142 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE6 [Vitis vinifera] |
| 3 |
Hb_001946_420 |
0.0196249481 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_092416_020 |
0.0231844527 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_005276_180 |
0.0259816576 |
- |
- |
hypothetical protein POPTR_0005s23040g [Populus trichocarpa] |
| 6 |
Hb_031546_010 |
0.0344657803 |
- |
- |
hypothetical protein JCGZ_21533 [Jatropha curcas] |
| 7 |
Hb_093084_040 |
0.0800650644 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0015s09370g [Populus trichocarpa] |
| 8 |
Hb_041188_010 |
0.082421882 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Nicotiana tomentosiformis] |
| 9 |
Hb_025526_110 |
0.0856812122 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 10 |
Hb_106677_010 |
0.0880487851 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 11 |
Hb_020712_020 |
0.0942905884 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Elaeis guineensis] |
| 12 |
Hb_005699_100 |
0.0943642133 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 13 |
Hb_000479_100 |
0.0998939414 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic [Gossypium arboreum] |
| 14 |
Hb_000355_020 |
0.1032646681 |
- |
- |
- |
| 15 |
Hb_001278_010 |
0.105565173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000645_040 |
0.1064640247 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 17 |
Hb_000002_190 |
0.11128164 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 18 |
Hb_000003_520 |
0.11128164 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 19 |
Hb_000005_170 |
0.11128164 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR [Jatropha curcas] |
| 20 |
Hb_000009_230 |
0.11128164 |
- |
- |
hypothetical protein POPTR_0007s12290g [Populus trichocarpa] |