| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
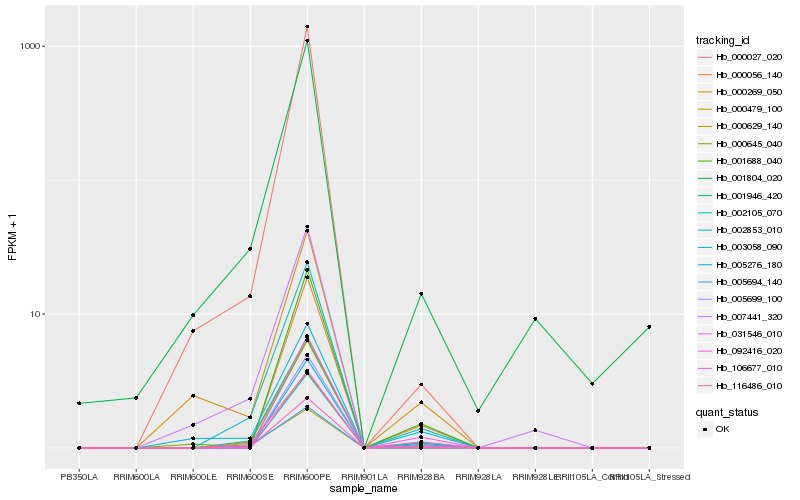
Hb_000479_100 |
0.0 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme, chloroplastic/amyloplastic [Gossypium arboreum] |
| 2 |
Hb_106677_010 |
0.0179021379 |
- |
- |
3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002853_010 |
0.0259925538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002105_070 |
0.0950223571 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005699_100 |
0.096491146 |
- |
- |
hypothetical protein POPTR_0013s12390g [Populus trichocarpa] |
| 6 |
Hb_116486_010 |
0.0983846582 |
- |
- |
MADS-box transcription factor, partial [Sapria himalayana] |
| 7 |
Hb_005276_180 |
0.0983958023 |
- |
- |
hypothetical protein POPTR_0005s23040g [Populus trichocarpa] |
| 8 |
Hb_000027_020 |
0.0988562466 |
- |
- |
Nonspecific lipid-transfer protein, putative [Ricinus communis] |
| 9 |
Hb_001688_040 |
0.0998939414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005694_140 |
0.1002327321 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000056_140 |
0.1005750527 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE6 [Vitis vinifera] |
| 12 |
Hb_001946_420 |
0.1052690967 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_092416_020 |
0.1065911953 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_031546_010 |
0.1114047099 |
- |
- |
hypothetical protein JCGZ_21533 [Jatropha curcas] |
| 15 |
Hb_000645_040 |
0.1146293195 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 16 |
Hb_001804_020 |
0.1162355697 |
- |
- |
hypothetical protein POPTR_0006s09110g [Populus trichocarpa] |
| 17 |
Hb_003058_090 |
0.1202480139 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 18 |
Hb_000629_140 |
0.1261020833 |
- |
- |
hypothetical protein SORBIDRAFT_04g013083 [Sorghum bicolor] |
| 19 |
Hb_007441_320 |
0.1292974026 |
- |
- |
PREDICTED: putative invertase inhibitor [Jatropha curcas] |
| 20 |
Hb_000269_050 |
0.1324129071 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |