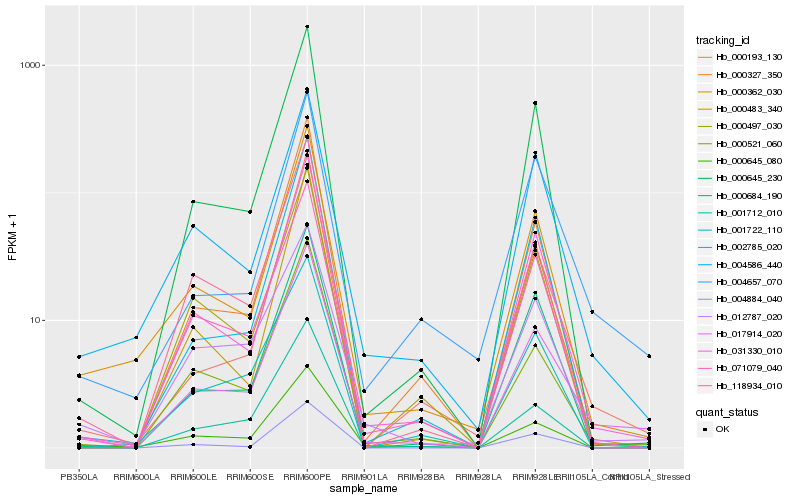
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017914_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_031330_010 |
0.0939000557 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000521_060 |
0.1058633728 |
- |
- |
PREDICTED: protein YLS3-like [Jatropha curcas] |
| 4 |
Hb_000362_030 |
0.1082387078 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 5 |
Hb_071079_040 |
0.1100198717 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET3 [Jatropha curcas] |
| 6 |
Hb_000645_230 |
0.1112401733 |
- |
- |
PREDICTED: chitinase-like protein 2 [Jatropha curcas] |
| 7 |
Hb_000645_080 |
0.1113913497 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 8 |
Hb_000193_130 |
0.1173849486 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_012787_020 |
0.120119108 |
- |
- |
PREDICTED: thioredoxin H7-like [Jatropha curcas] |
| 10 |
Hb_118934_010 |
0.1209742577 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_002785_020 |
0.1233844835 |
- |
- |
PREDICTED: laccase-4-like [Jatropha curcas] |
| 12 |
Hb_004586_440 |
0.1238106322 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 13 |
Hb_000327_350 |
0.1240162301 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 14 |
Hb_004657_070 |
0.1248728885 |
- |
- |
PREDICTED: protein IRX15-LIKE [Jatropha curcas] |
| 15 |
Hb_000684_190 |
0.1267038079 |
- |
- |
beta-1,3-glucuronyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000483_340 |
0.1275604432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001712_010 |
0.1283463097 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 1 [Jatropha curcas] |
| 18 |
Hb_004884_040 |
0.1290999495 |
- |
- |
hypothetical protein POPTR_0016s12910g [Populus trichocarpa] |
| 19 |
Hb_001722_110 |
0.1294375297 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_000497_030 |
0.1295980334 |
- |
- |
- |