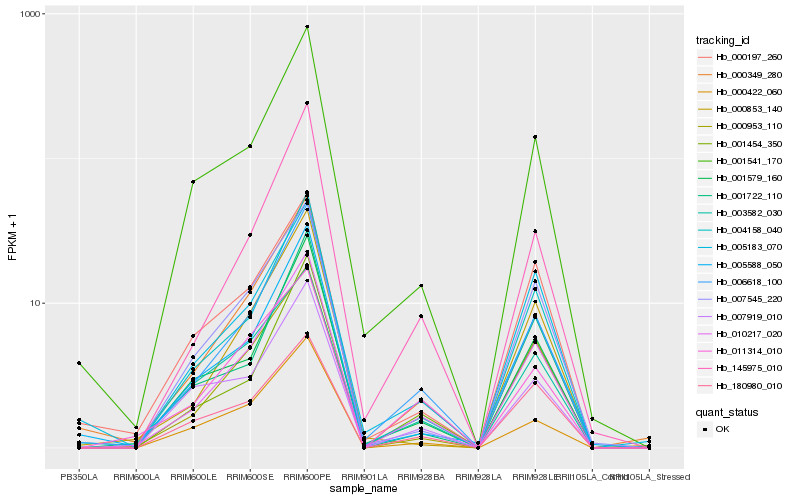
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010217_020 |
0.0 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 2 |
Hb_006618_100 |
0.0599712843 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 3 |
Hb_000953_110 |
0.0742537663 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 4 |
Hb_004158_040 |
0.0774428517 |
- |
- |
PREDICTED: WAT1-related protein At2g37460-like [Jatropha curcas] |
| 5 |
Hb_001454_350 |
0.0870436181 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 6 |
Hb_000349_280 |
0.0871361163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_007545_220 |
0.0956442671 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 8 |
Hb_001579_160 |
0.0958433691 |
- |
- |
PREDICTED: kininogen-1-like [Jatropha curcas] |
| 9 |
Hb_145975_010 |
0.0991989591 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_005588_050 |
0.1022362642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001541_170 |
0.1033470484 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 12 |
Hb_000422_060 |
0.1067590889 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_1282480 [Ricinus communis] |
| 13 |
Hb_000853_140 |
0.1078052021 |
- |
- |
hypothetical protein POPTR_0014s12080g [Populus trichocarpa] |
| 14 |
Hb_003582_030 |
0.1089945904 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 15 |
Hb_005183_070 |
0.1103836199 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 16 |
Hb_001722_110 |
0.1109051895 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_180980_010 |
0.1154466515 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 18 |
Hb_000197_260 |
0.1179551991 |
- |
- |
hypothetical protein PRUPE_ppa025448mg [Prunus persica] |
| 19 |
Hb_011314_010 |
0.1187897102 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_007919_010 |
0.1194629089 |
- |
- |
conserved hypothetical protein [Ricinus communis] |