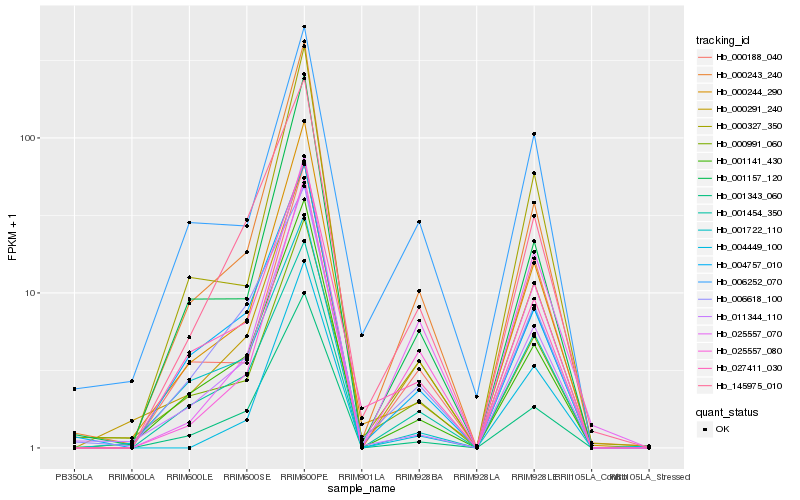
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_240 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s16430g [Populus trichocarpa] |
| 2 |
Hb_000991_060 |
0.0862070663 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 3 |
Hb_025557_080 |
0.0909546862 |
transcription factor |
TF Family: zf-HD |
- |
| 4 |
Hb_001454_350 |
0.0941800641 |
- |
- |
PREDICTED: uncharacterized protein LOC105643449 [Jatropha curcas] |
| 5 |
Hb_000243_240 |
0.0993180922 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 6 |
Hb_145975_010 |
0.1019375023 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_001141_430 |
0.1022107781 |
- |
- |
hypothetical protein POPTR_0008s15160g [Populus trichocarpa] |
| 8 |
Hb_000188_040 |
0.1028627039 |
- |
- |
PREDICTED: uncharacterized protein LOC105629522 [Jatropha curcas] |
| 9 |
Hb_006252_070 |
0.1098514711 |
- |
- |
hypothetical protein JCGZ_17254 [Jatropha curcas] |
| 10 |
Hb_000244_290 |
0.1098898537 |
- |
- |
PREDICTED: UDP-glucuronate:xylan alpha-glucuronosyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_001343_060 |
0.1161108795 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 12 |
Hb_027411_030 |
0.1174595361 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 13 |
Hb_011344_110 |
0.1181279836 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 14 |
Hb_004757_010 |
0.1196379152 |
- |
- |
PREDICTED: uncharacterized protein LOC105121200 [Populus euphratica] |
| 15 |
Hb_025557_070 |
0.1202778687 |
transcription factor |
TF Family: zf-HD |
hypothetical protein JCGZ_11626 [Jatropha curcas] |
| 16 |
Hb_001157_120 |
0.123596172 |
- |
- |
PREDICTED: dirigent protein 19-like [Jatropha curcas] |
| 17 |
Hb_000327_350 |
0.1238630926 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 18 |
Hb_006618_100 |
0.1251781146 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus tremula x Populus tremuloides] |
| 19 |
Hb_004449_100 |
0.1264108668 |
- |
- |
- |
| 20 |
Hb_001722_110 |
0.1265346997 |
- |
- |
DUF26 domain-containing protein 1 precursor, putative [Ricinus communis] |