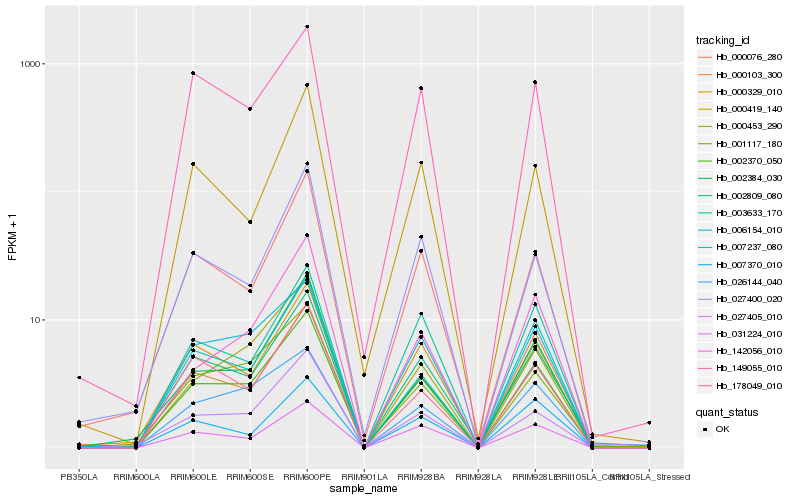
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002370_050 |
0.0 |
- |
- |
hypothetical protein JCGZ_18849 [Jatropha curcas] |
| 2 |
Hb_007237_080 |
0.0813205021 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_002809_080 |
0.0823573955 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_031224_010 |
0.0873534252 |
- |
- |
- |
| 5 |
Hb_000329_010 |
0.094085172 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 6 |
Hb_027405_010 |
0.0971713446 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 7 |
Hb_026144_040 |
0.1002626972 |
- |
- |
- |
| 8 |
Hb_003633_170 |
0.1008163248 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000453_290 |
0.1009426349 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 10 |
Hb_006154_010 |
0.1026652294 |
- |
- |
UDP-glucose 6-dehydrogenase, putative [Ricinus communis] |
| 11 |
Hb_000076_280 |
0.1039241623 |
- |
- |
PREDICTED: pectinesterase-like [Populus euphratica] |
| 12 |
Hb_001117_180 |
0.1077024982 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 13 |
Hb_002384_030 |
0.1121447526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_149055_010 |
0.1127492766 |
- |
- |
cinnamate 4-hydroxylase [Leucaena leucocephala] |
| 15 |
Hb_178049_010 |
0.1140654415 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000419_140 |
0.1177129416 |
- |
- |
Aquaporin PIP2.1, putative [Ricinus communis] |
| 17 |
Hb_000103_300 |
0.1178411124 |
- |
- |
PREDICTED: perakine reductase-like [Jatropha curcas] |
| 18 |
Hb_142056_010 |
0.1194556443 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007370_010 |
0.120060339 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 20 |
Hb_027400_020 |
0.1211163515 |
- |
- |
PREDICTED: uncharacterized protein At5g22580 isoform X2 [Jatropha curcas] |