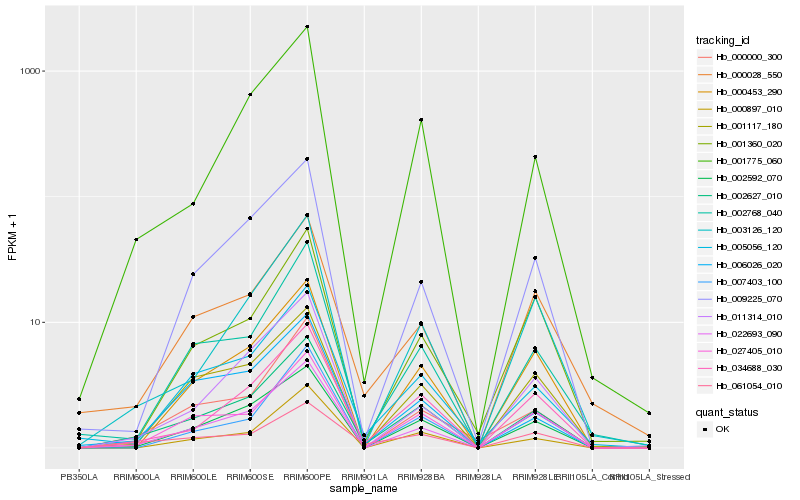
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002627_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640888 [Jatropha curcas] |
| 2 |
Hb_034688_030 |
0.0943091705 |
- |
- |
PREDICTED: uncharacterized protein LOC105642049 [Jatropha curcas] |
| 3 |
Hb_002592_070 |
0.1034191529 |
- |
- |
hypothetical protein VITISV_020993 [Vitis vinifera] |
| 4 |
Hb_027405_010 |
0.1065090652 |
- |
- |
PREDICTED: uncharacterized protein At3g17950-like [Populus euphratica] |
| 5 |
Hb_022693_090 |
0.1131458711 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_061054_010 |
0.119112617 |
- |
- |
hypothetical protein JCGZ_01675 [Jatropha curcas] |
| 7 |
Hb_001775_060 |
0.1199981131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005056_120 |
0.121352192 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_000000_300 |
0.1258359118 |
- |
- |
PREDICTED: pathogenesis-related protein 5 [Jatropha curcas] |
| 10 |
Hb_002768_040 |
0.1258737615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000453_290 |
0.1285843185 |
- |
- |
PREDICTED: dr1-associated corepressor-like [Jatropha curcas] |
| 12 |
Hb_001117_180 |
0.1297687963 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003126_120 |
0.1313974017 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Jatropha curcas] |
| 14 |
Hb_009225_070 |
0.1359282669 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 15 |
Hb_000028_550 |
0.1377628697 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_000897_010 |
0.1386626563 |
- |
- |
hypothetical protein JCGZ_13196 [Jatropha curcas] |
| 17 |
Hb_011314_010 |
0.1397727846 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_006026_020 |
0.1457008434 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g48800 [Jatropha curcas] |
| 19 |
Hb_007403_100 |
0.1480947305 |
- |
- |
PREDICTED: probable purple acid phosphatase 20 [Jatropha curcas] |
| 20 |
Hb_001360_020 |
0.1497352559 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 [Nelumbo nucifera] |