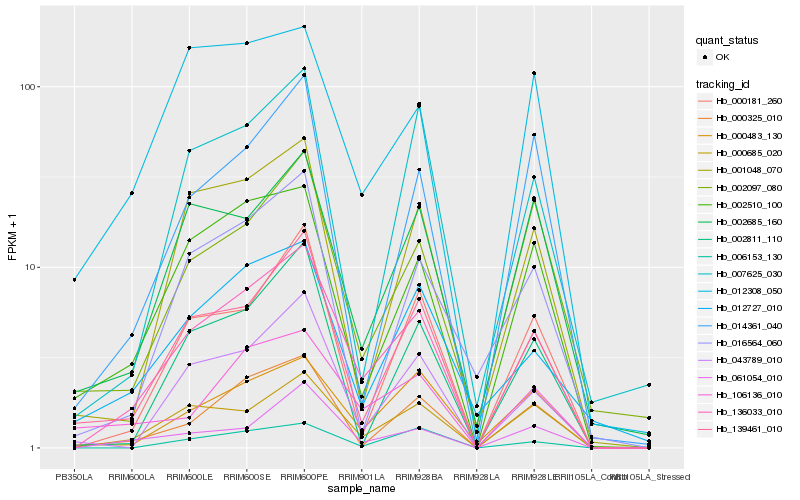
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136033_010 |
0.0 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Gossypium raimondii] |
| 2 |
Hb_000685_020 |
0.1243514433 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 4-like [Jatropha curcas] |
| 3 |
Hb_139461_010 |
0.1360559115 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 4 |
Hb_000325_010 |
0.1499081083 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 5 |
Hb_002510_100 |
0.1519070124 |
- |
- |
Root phototropism protein, putative [Ricinus communis] |
| 6 |
Hb_043789_010 |
0.1520733314 |
transcription factor |
TF Family: AP2 |
hypothetical protein VITISV_034131 [Vitis vinifera] |
| 7 |
Hb_061054_010 |
0.1534177991 |
- |
- |
hypothetical protein JCGZ_01675 [Jatropha curcas] |
| 8 |
Hb_000181_260 |
0.1555181388 |
- |
- |
PREDICTED: uncharacterized protein LOC105139156 [Populus euphratica] |
| 9 |
Hb_000483_130 |
0.1557568107 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001048_070 |
0.1641690066 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 11 |
Hb_106136_010 |
0.1667848253 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_002685_160 |
0.1691811667 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 13 |
Hb_006153_130 |
0.171448702 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002811_110 |
0.1719956979 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 15 |
Hb_012727_010 |
0.1726623064 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 16 |
Hb_014361_040 |
0.1748297381 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 17 |
Hb_007625_030 |
0.1764673338 |
- |
- |
unknown [Medicago truncatula] |
| 18 |
Hb_016564_060 |
0.1767948057 |
- |
- |
PREDICTED: uncharacterized protein LOC105628249 [Jatropha curcas] |
| 19 |
Hb_012308_050 |
0.1777073695 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA9-like [Jatropha curcas] |
| 20 |
Hb_002097_080 |
0.1778119728 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |