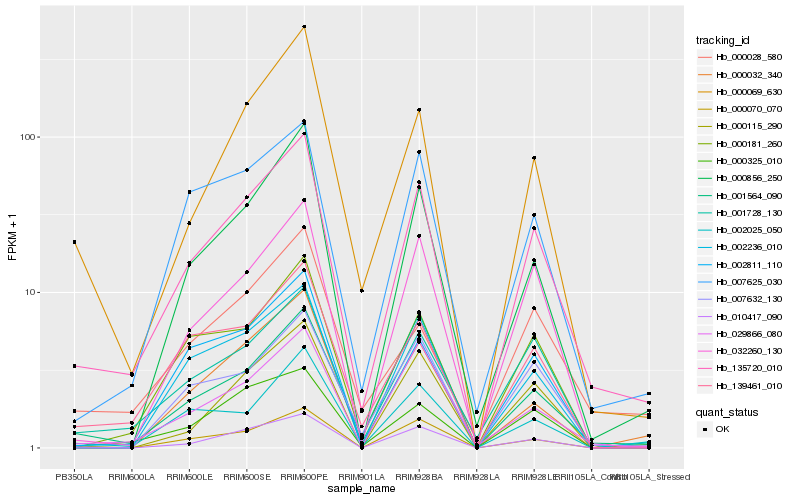
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135720_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_17857 [Jatropha curcas] |
| 2 |
Hb_001728_130 |
0.1173187152 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_007625_030 |
0.1267914593 |
- |
- |
unknown [Medicago truncatula] |
| 4 |
Hb_029866_080 |
0.1269884146 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 5 |
Hb_032260_130 |
0.1292560543 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 6 |
Hb_000325_010 |
0.131025316 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 7 |
Hb_139461_010 |
0.1322334748 |
desease resistance |
Gene Name: NB-ARC |
Cc-nbs-lrr resistance protein, putative [Theobroma cacao] |
| 8 |
Hb_000181_260 |
0.1379831918 |
- |
- |
PREDICTED: uncharacterized protein LOC105139156 [Populus euphratica] |
| 9 |
Hb_000856_250 |
0.138726085 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000115_290 |
0.1395274415 |
- |
- |
PREDICTED: uncharacterized protein LOC105124123 [Populus euphratica] |
| 11 |
Hb_000069_630 |
0.1396411511 |
- |
- |
hypothetical protein [Hevea brasiliensis] |
| 12 |
Hb_010417_090 |
0.1406921322 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 13 |
Hb_007632_130 |
0.1408790663 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001564_090 |
0.1432144607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002025_050 |
0.1436343453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002236_010 |
0.1449963851 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4 [Jatropha curcas] |
| 17 |
Hb_002811_110 |
0.1451066942 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 18 |
Hb_000070_070 |
0.1501318807 |
- |
- |
PREDICTED: protein LURP-one-related 17 [Jatropha curcas] |
| 19 |
Hb_000032_340 |
0.1503622963 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 20 |
Hb_000028_580 |
0.1531827885 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |