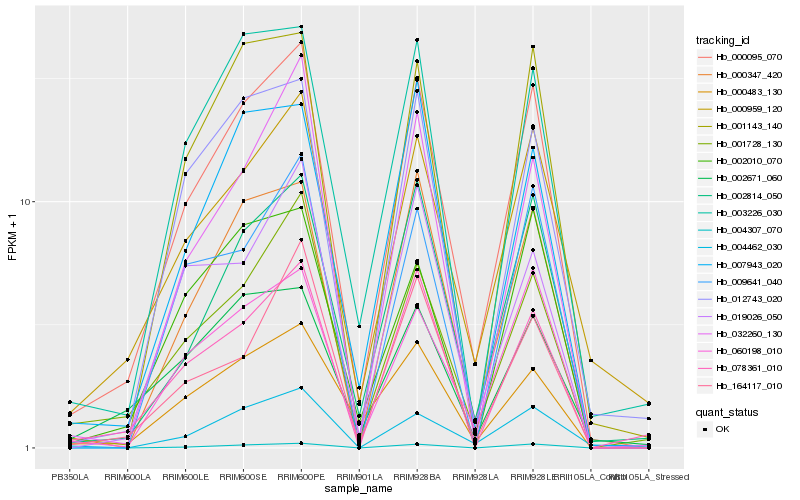
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000095_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002814_050 |
0.0978976318 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 3 |
Hb_060198_010 |
0.1029495173 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 4 |
Hb_000347_420 |
0.107990956 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 5 |
Hb_001728_130 |
0.1119694065 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000959_120 |
0.1122690625 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 7 |
Hb_004307_070 |
0.1136218128 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_003226_030 |
0.1192552491 |
- |
- |
sucrose synthase 5 [Hevea brasiliensis] |
| 9 |
Hb_001143_140 |
0.1202165024 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_012743_020 |
0.1236407732 |
- |
- |
serine/threonine protein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_000483_130 |
0.1247252903 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 12 |
Hb_007943_020 |
0.1278199839 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_078361_010 |
0.1279770471 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 14 |
Hb_004462_030 |
0.1304334123 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 15 |
Hb_032260_130 |
0.1314180685 |
- |
- |
hypothetical protein POPTR_0003s13800g [Populus trichocarpa] |
| 16 |
Hb_164117_010 |
0.1366744487 |
- |
- |
hypothetical protein PHAVU_004G0480000g [Phaseolus vulgaris] |
| 17 |
Hb_002010_070 |
0.1385051606 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 18 |
Hb_002671_060 |
0.1397907139 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 19 |
Hb_009641_040 |
0.1401902266 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 20 |
Hb_019026_050 |
0.1402265213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |