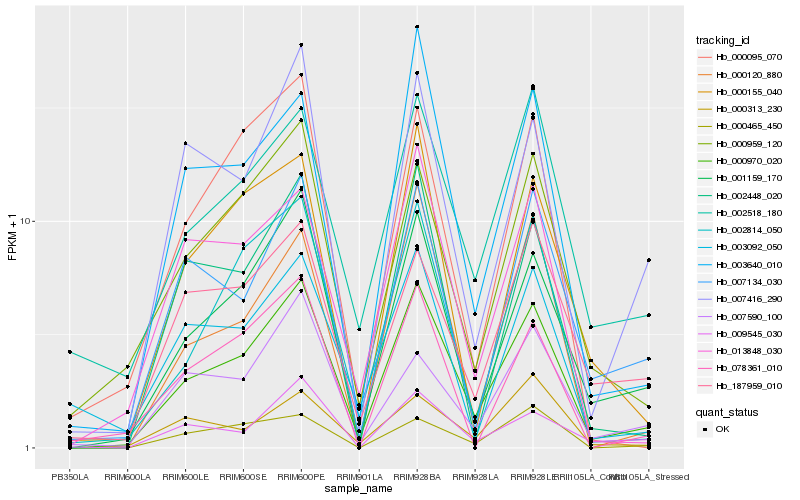
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000970_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 2 |
Hb_001159_170 |
0.0992846125 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 3 |
Hb_000959_120 |
0.1328547615 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 4 |
Hb_002814_050 |
0.1331397723 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 5 |
Hb_003640_010 |
0.1368065279 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 6 |
Hb_002518_180 |
0.1386644387 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 7 |
Hb_007416_290 |
0.1394616526 |
- |
- |
PREDICTED: plasma membrane ATPase 4-like [Pyrus x bretschneideri] |
| 8 |
Hb_007134_030 |
0.1406346681 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 9 |
Hb_013848_030 |
0.1452183123 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 10 |
Hb_000120_880 |
0.1467967139 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 11 |
Hb_002448_020 |
0.1508019417 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 12 |
Hb_187959_010 |
0.1513626801 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 13 |
Hb_000155_040 |
0.1544020158 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 14 |
Hb_009545_030 |
0.1549025401 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000095_070 |
0.1552364176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007590_100 |
0.1584159087 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 17 |
Hb_000313_230 |
0.1614295763 |
- |
- |
PREDICTED: uncharacterized protein LOC105636893 [Jatropha curcas] |
| 18 |
Hb_000465_450 |
0.161691202 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 19 |
Hb_003092_050 |
0.164123384 |
- |
- |
PREDICTED: putative chloride channel-like protein CLC-g [Jatropha curcas] |
| 20 |
Hb_078361_010 |
0.1673153405 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |