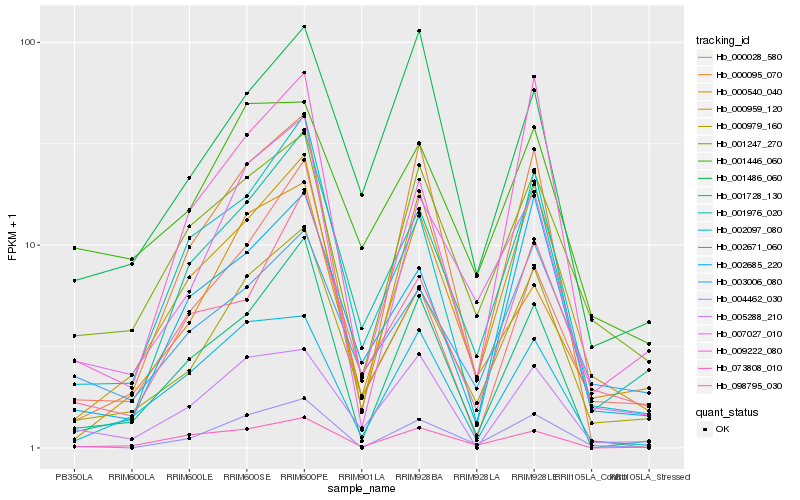
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_160 |
0.0 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 2 |
Hb_001976_020 |
0.0993680522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000959_120 |
0.1142910825 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 4 |
Hb_007027_010 |
0.1263510012 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 5 |
Hb_002097_080 |
0.1334790297 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 6 |
Hb_001247_270 |
0.1336139677 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003006_080 |
0.133773501 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_098795_030 |
0.135906652 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 9 |
Hb_002685_220 |
0.1385437796 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_004462_030 |
0.1501620876 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 11 |
Hb_001486_060 |
0.1529194801 |
- |
- |
PREDICTED: uncharacterized protein LOC105632622 [Jatropha curcas] |
| 12 |
Hb_001446_060 |
0.1539508666 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_001728_130 |
0.1539792356 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_073808_010 |
0.1589969001 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000095_070 |
0.1631119271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005288_210 |
0.1631618608 |
- |
- |
hypothetical protein VITISV_017217 [Vitis vinifera] |
| 17 |
Hb_000540_040 |
0.163564015 |
- |
- |
PREDICTED: chitinase 2 [Amborella trichopoda] |
| 18 |
Hb_009222_080 |
0.165613646 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |
| 19 |
Hb_002671_060 |
0.1664741047 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 20 |
Hb_000028_580 |
0.1675392549 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |