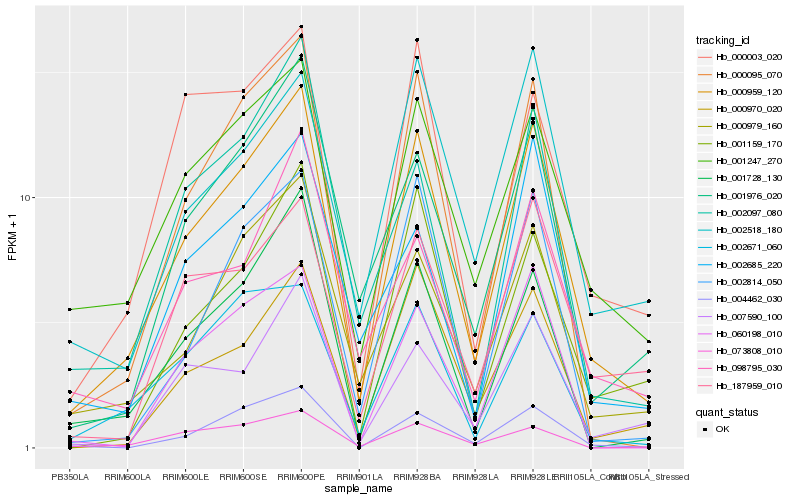
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000959_120 |
0.0 |
- |
- |
PREDICTED: protein BRASSINOSTEROID INSENSITIVE 1 [Jatropha curcas] |
| 2 |
Hb_000095_070 |
0.1122690625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001247_270 |
0.1136694678 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000979_160 |
0.1142910825 |
- |
- |
hypothetical protein RCOM_0629030 [Ricinus communis] |
| 5 |
Hb_001728_130 |
0.1285285499 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001976_020 |
0.1320730194 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004462_030 |
0.1324395303 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 8 |
Hb_000970_020 |
0.1328547615 |
- |
- |
hypothetical protein POPTR_0005s19050g [Populus trichocarpa] |
| 9 |
Hb_187959_010 |
0.135679535 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 10 |
Hb_002671_060 |
0.1359614286 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 11 |
Hb_001159_170 |
0.1359986403 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1 [Jatropha curcas] |
| 12 |
Hb_073808_010 |
0.1368462301 |
desease resistance |
Gene Name: DUF594 |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000003_020 |
0.1375248405 |
- |
- |
PREDICTED: uncharacterized protein LOC105631161 [Jatropha curcas] |
| 14 |
Hb_002814_050 |
0.1405884862 |
- |
- |
PREDICTED: sugar carrier protein A [Jatropha curcas] |
| 15 |
Hb_007590_100 |
0.1427196368 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 16 |
Hb_002097_080 |
0.1427300798 |
- |
- |
phosphatidylinositol-4-phosphate 5-kinase, putative [Ricinus communis] |
| 17 |
Hb_098795_030 |
0.145917071 |
- |
- |
PREDICTED: metal tolerance protein 1-like [Vitis vinifera] |
| 18 |
Hb_002685_220 |
0.147022127 |
- |
- |
PREDICTED: cationic amino acid transporter 4, vacuolar-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002518_180 |
0.1502845603 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 20 |
Hb_060198_010 |
0.1502921253 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |