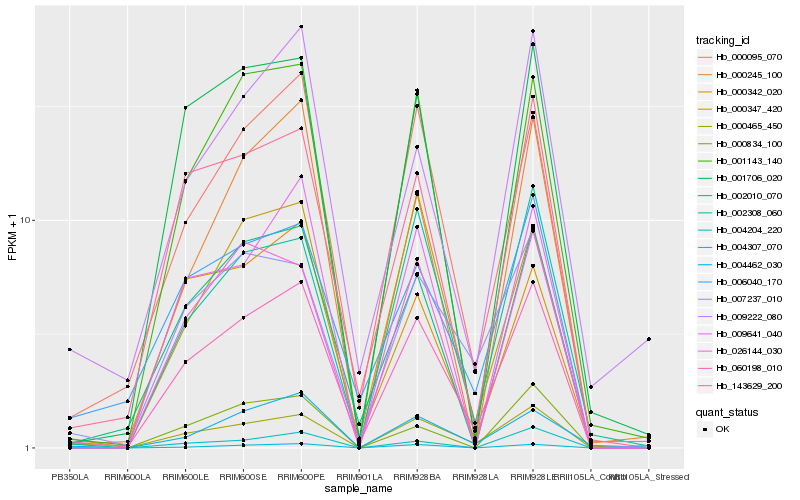
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_060198_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 2 |
Hb_000095_070 |
0.1029495173 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002010_070 |
0.1139904435 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |
| 4 |
Hb_143629_200 |
0.1148054735 |
- |
- |
hypothetical protein POPTR_0013s00300g [Populus trichocarpa] |
| 5 |
Hb_001143_140 |
0.1191606377 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_000465_450 |
0.1216935594 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 7 |
Hb_001706_020 |
0.1234784478 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 8 |
Hb_006040_170 |
0.1269801429 |
- |
- |
NADH-ubiquinone oxidoreductase chain [Medicago truncatula] |
| 9 |
Hb_009641_040 |
0.1282686462 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 10 |
Hb_004307_070 |
0.1309275112 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_007237_010 |
0.1311393444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000245_100 |
0.1372890932 |
- |
- |
PREDICTED: uncharacterized protein LOC105631555 [Jatropha curcas] |
| 13 |
Hb_002308_060 |
0.1397814821 |
- |
- |
PREDICTED: cytochrome P450 704C1-like [Jatropha curcas] |
| 14 |
Hb_004462_030 |
0.1399715566 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 15 |
Hb_000342_020 |
0.141324475 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 16 |
Hb_000834_100 |
0.1436849277 |
- |
- |
PREDICTED: uncharacterized protein LOC105178355 [Sesamum indicum] |
| 17 |
Hb_000347_420 |
0.1459581315 |
- |
- |
hypothetical protein JCGZ_24696 [Jatropha curcas] |
| 18 |
Hb_026144_030 |
0.1474926099 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 19 |
Hb_004204_220 |
0.1476659731 |
- |
- |
- |
| 20 |
Hb_009222_080 |
0.1484056913 |
- |
- |
RING-H2 finger protein ATL2J, putative [Ricinus communis] |