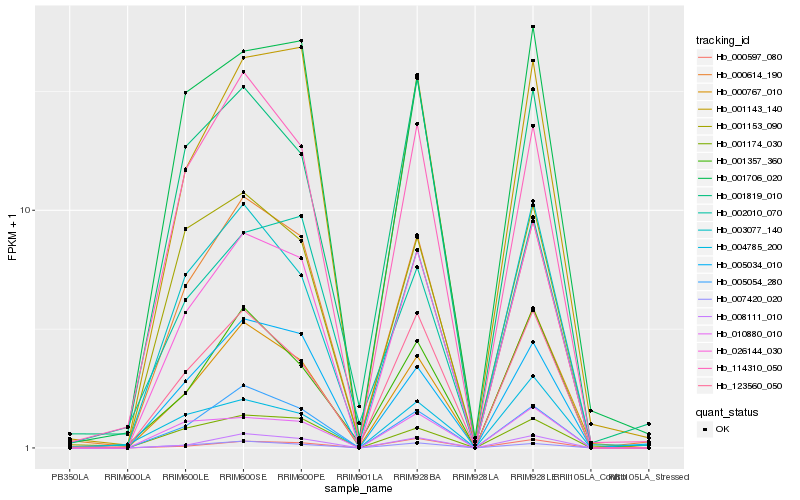
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026144_030 |
0.0 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 2 |
Hb_008111_010 |
0.0797044119 |
- |
- |
hypothetical protein VITISV_021527 [Vitis vinifera] |
| 3 |
Hb_000614_190 |
0.0840074156 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 4 |
Hb_000767_010 |
0.0896615662 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 5 |
Hb_001143_140 |
0.0947921898 |
- |
- |
Aldehyde dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_123560_050 |
0.0957488423 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 7 |
Hb_007420_020 |
0.0961828951 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_003077_140 |
0.0985569625 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 9 |
Hb_010880_010 |
0.0988020543 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005054_280 |
0.0993360162 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 11 |
Hb_001706_020 |
0.1000069995 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 12 |
Hb_001174_030 |
0.1019114166 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000597_080 |
0.1156811383 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_001357_360 |
0.1163259501 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_005034_010 |
0.1179811656 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_004785_200 |
0.1200035327 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 17 |
Hb_114310_050 |
0.1200951336 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 18 |
Hb_001819_010 |
0.1245256961 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 19 |
Hb_001153_090 |
0.1248169288 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 20 |
Hb_002010_070 |
0.1310676242 |
- |
- |
hypothetical protein EUGRSUZ_I027342, partial [Eucalyptus grandis] |