| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
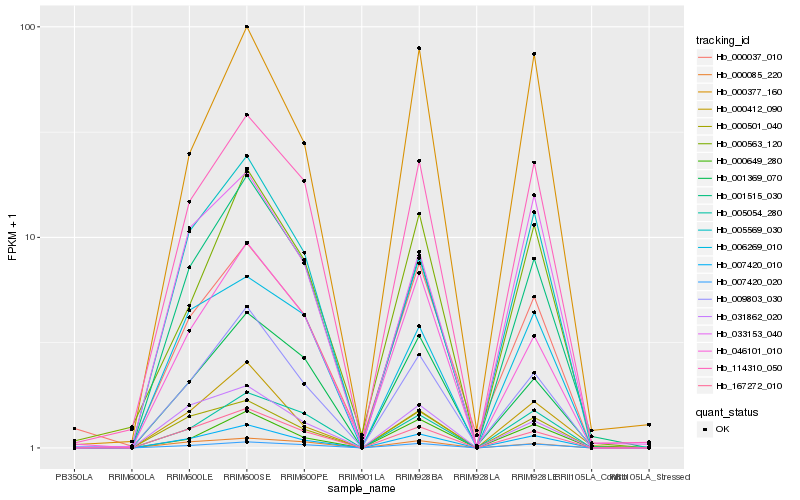
| 1 |
Hb_007420_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X2 [Jatropha curcas] |
| 2 |
Hb_167272_010 |
0.0686576395 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 3 |
Hb_000501_040 |
0.0700887845 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_114310_050 |
0.0761221622 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 5 |
Hb_007420_020 |
0.0781825096 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 6 |
Hb_009803_030 |
0.0800237157 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 7 |
Hb_001515_030 |
0.0882854437 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_031862_020 |
0.1021393132 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 9 |
Hb_000649_280 |
0.1060244018 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 10 |
Hb_005054_280 |
0.1062258456 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 11 |
Hb_005569_030 |
0.1086207496 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000037_010 |
0.1128767448 |
- |
- |
PREDICTED: cation/H(+) antiporter 18-like [Jatropha curcas] |
| 13 |
Hb_000412_090 |
0.1141183868 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 14 |
Hb_046101_010 |
0.1143178098 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase-like protein CCR3 [Jatropha curcas] |
| 15 |
Hb_000377_160 |
0.1163021352 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 16 |
Hb_000563_120 |
0.116886664 |
- |
- |
hypothetical protein JCGZ_18936 [Jatropha curcas] |
| 17 |
Hb_033153_040 |
0.1186515413 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 18 |
Hb_001369_070 |
0.1193500756 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000085_220 |
0.1204164435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006269_010 |
0.1207158545 |
- |
- |
hypothetical protein JCGZ_21552 [Jatropha curcas] |