| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
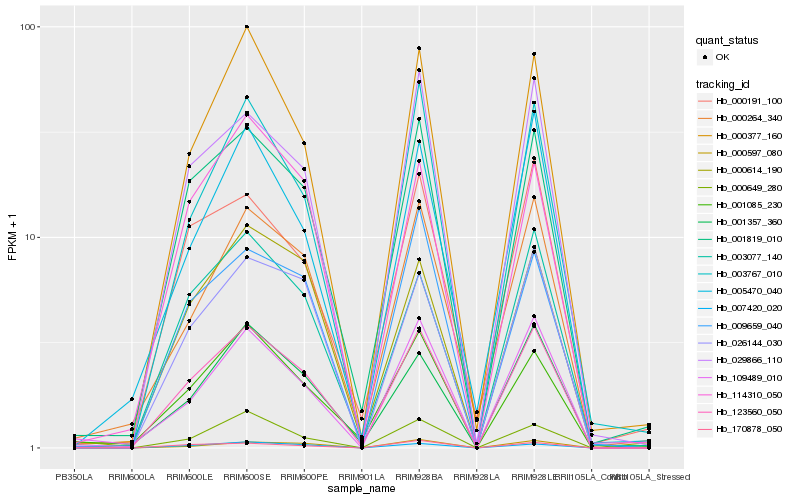
Hb_123560_050 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 2 |
Hb_001819_010 |
0.0614210156 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 3 |
Hb_000377_160 |
0.0854020645 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 4 |
Hb_003077_140 |
0.0924958951 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 5 |
Hb_029866_110 |
0.0930805415 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 10-like [Jatropha curcas] |
| 6 |
Hb_007420_020 |
0.094770278 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_026144_030 |
0.0957488423 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 8 |
Hb_001357_360 |
0.1010852344 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_003767_010 |
0.101347629 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD11 [Jatropha curcas] |
| 10 |
Hb_001085_230 |
0.10319753 |
- |
- |
PREDICTED: receptor like protein kinase S.2 [Jatropha curcas] |
| 11 |
Hb_000614_190 |
0.1049362339 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 12 |
Hb_000264_340 |
0.1078858544 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_109489_010 |
0.109917725 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 14 |
Hb_114310_050 |
0.1108238274 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 15 |
Hb_009659_040 |
0.1108365132 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 16 |
Hb_170878_050 |
0.1117840418 |
- |
- |
retrotransposon protein, putative, unclassified [Oryza sativa Japonica Group] |
| 17 |
Hb_000649_280 |
0.1124490116 |
- |
- |
PREDICTED: F-box protein PP2-B10-like [Pyrus x bretschneideri] |
| 18 |
Hb_005470_040 |
0.113878776 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 19 |
Hb_000191_100 |
0.114087134 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_000597_080 |
0.1160664076 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like isoform X2 [Jatropha curcas] |