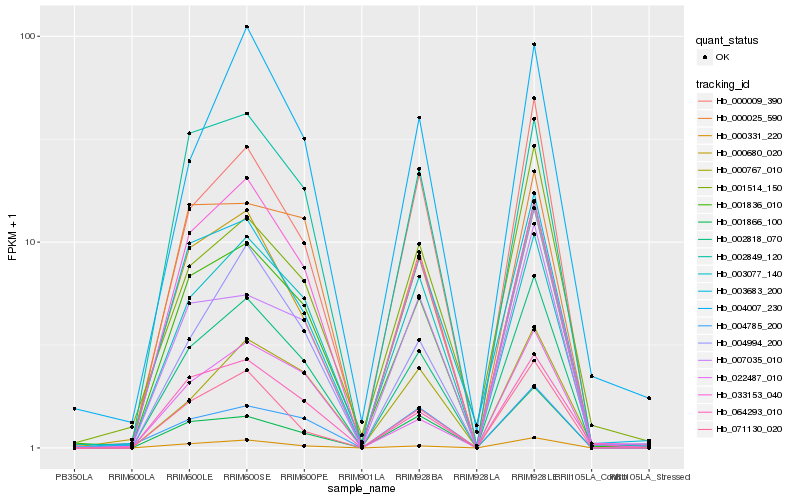
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002818_070 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001836_010 |
0.0661723488 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 3 |
Hb_000009_390 |
0.0869531308 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000331_220 |
0.0873887633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003077_140 |
0.0955488464 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 6 |
Hb_071130_020 |
0.0985876109 |
- |
- |
- |
| 7 |
Hb_000680_020 |
0.1089740408 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_003683_200 |
0.112098994 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 9 |
Hb_033153_040 |
0.1138735688 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_004994_200 |
0.1165496584 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 11 |
Hb_000767_010 |
0.1170252116 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 12 |
Hb_007035_010 |
0.117473278 |
- |
- |
PREDICTED: synaptotagmin-4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_064293_010 |
0.1181879311 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 14 |
Hb_001866_100 |
0.1192292684 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 15 |
Hb_002849_120 |
0.1285054389 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 16 |
Hb_004785_200 |
0.1298022511 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 17 |
Hb_022487_010 |
0.1335992112 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 18 |
Hb_004007_230 |
0.1337177343 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000025_590 |
0.1355835454 |
- |
- |
PREDICTED: cytochrome P450 94A1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001514_150 |
0.1366945633 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |