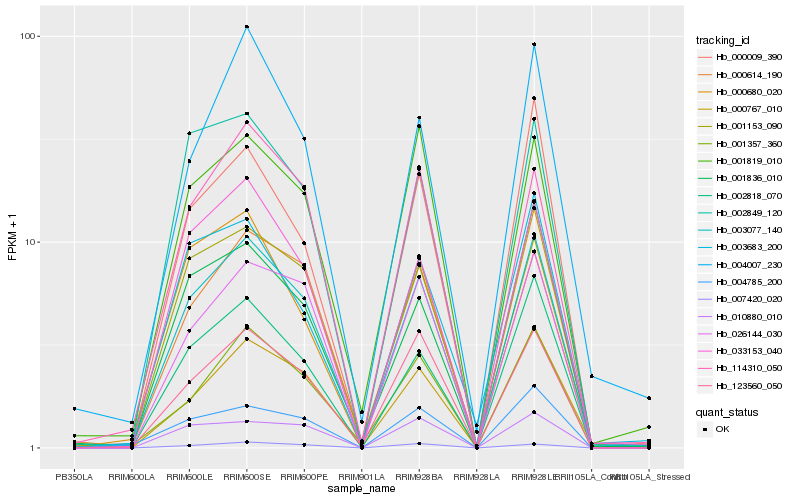
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003077_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 2 |
Hb_000767_010 |
0.0880446683 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 3 |
Hb_001153_090 |
0.0882648715 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 4 |
Hb_033153_040 |
0.0894949098 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_123560_050 |
0.0924958951 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 [Jatropha curcas] |
| 6 |
Hb_001357_360 |
0.0945978735 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_002818_070 |
0.0955488464 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_026144_030 |
0.0985569625 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.5 [Jatropha curcas] |
| 9 |
Hb_000614_190 |
0.0993268996 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 10 |
Hb_000680_020 |
0.102291163 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_114310_050 |
0.1041484943 |
- |
- |
Iron(III)-zinc(II) purple acid phosphatase precursor family protein [Populus trichocarpa] |
| 12 |
Hb_002849_120 |
0.1062080264 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 13 |
Hb_001819_010 |
0.1092484861 |
- |
- |
PREDICTED: potassium transporter 8 [Jatropha curcas] |
| 14 |
Hb_004785_200 |
0.1113325594 |
- |
- |
PREDICTED: protein MKS1 [Jatropha curcas] |
| 15 |
Hb_001836_010 |
0.1121490172 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 16 |
Hb_004007_230 |
0.1139358222 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 17 |
Hb_007420_020 |
0.1146208938 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 18 |
Hb_010880_010 |
0.1196595114 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000009_390 |
0.121396965 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003683_200 |
0.1215274126 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |