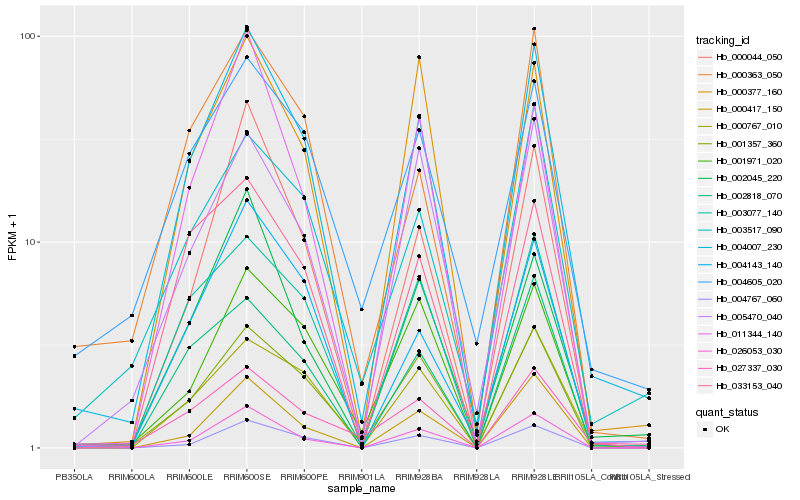
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004007_230 |
0.0 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 2 |
Hb_026053_030 |
0.1037116108 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 3 |
Hb_001357_360 |
0.109728686 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_004143_140 |
0.1113816419 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 5 |
Hb_000044_050 |
0.1137156139 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_003077_140 |
0.1139358222 |
- |
- |
hypothetical protein JCGZ_16294 [Jatropha curcas] |
| 7 |
Hb_000767_010 |
0.1161663508 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like isoform X2 [Solanum lycopersicum] |
| 8 |
Hb_000417_150 |
0.1174745909 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 9 |
Hb_002045_220 |
0.1223710645 |
- |
- |
PREDICTED: uncharacterized protein LOC105649919 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000363_050 |
0.1276097854 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_033153_040 |
0.1336404311 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 12 |
Hb_002818_070 |
0.1337177343 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_003517_090 |
0.1360552936 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_027337_030 |
0.1402000957 |
- |
- |
PREDICTED: uncharacterized protein LOC104602841 [Nelumbo nucifera] |
| 15 |
Hb_001971_020 |
0.1418928763 |
- |
- |
PREDICTED: alpha-dioxygenase 2 [Populus euphratica] |
| 16 |
Hb_005470_040 |
0.1437575241 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 17 |
Hb_000377_160 |
0.1447225325 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 18 |
Hb_004605_020 |
0.1454720292 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 19 |
Hb_011344_140 |
0.1463762486 |
- |
- |
PREDICTED: geraniol 8-hydroxylase-like [Jatropha curcas] |
| 20 |
Hb_004767_060 |
0.1465183797 |
- |
- |
PREDICTED: uncharacterized protein LOC105761928 [Gossypium raimondii] |