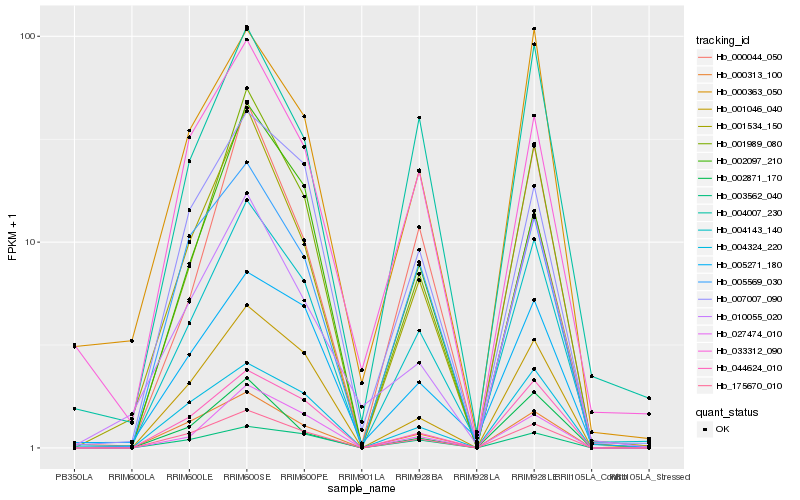
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004143_140 |
0.0 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 2 |
Hb_001989_080 |
0.0901605203 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_175670_010 |
0.0924854851 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 4 |
Hb_001046_040 |
0.0986555183 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_044624_010 |
0.1022706615 |
- |
- |
- |
| 6 |
Hb_027474_010 |
0.110582926 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 7 |
Hb_005271_180 |
0.1112707139 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 8 |
Hb_004007_230 |
0.1113816419 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000313_100 |
0.114114462 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 10 |
Hb_004324_220 |
0.1212782203 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 11 |
Hb_003562_040 |
0.1258322363 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 12 |
Hb_000363_050 |
0.1260990841 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000044_050 |
0.1265254239 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_007007_090 |
0.1275243256 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 15 |
Hb_005569_030 |
0.134985493 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 16 |
Hb_033312_090 |
0.1369724427 |
- |
- |
PREDICTED: HMG-Y-related protein A-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002097_210 |
0.1440625749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010055_020 |
0.146562993 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 19 |
Hb_001534_150 |
0.1469535381 |
- |
- |
PREDICTED: uncharacterized permease C29B12.14c [Jatropha curcas] |
| 20 |
Hb_002871_170 |
0.1489267976 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |