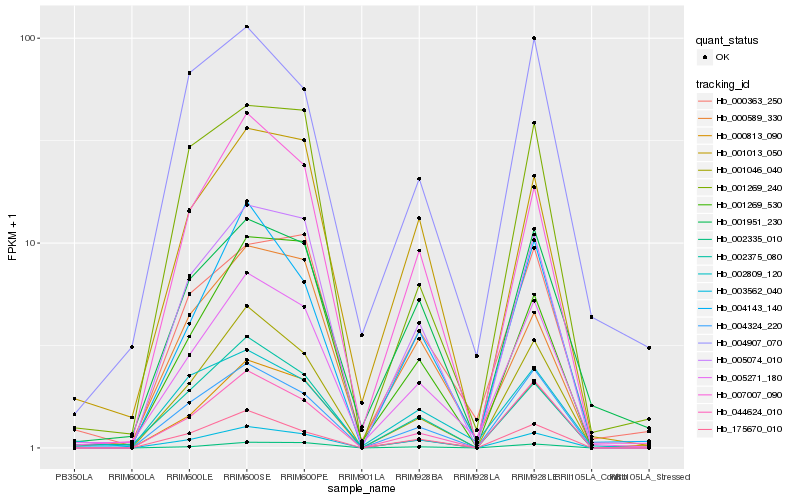
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005271_180 |
0.0 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 2 |
Hb_007007_090 |
0.1094133936 |
- |
- |
phospholipase d beta, putative [Ricinus communis] |
| 3 |
Hb_004143_140 |
0.1112707139 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 4 |
Hb_005074_010 |
0.1164314648 |
- |
- |
PREDICTED: uncharacterized protein LOC105644456 [Jatropha curcas] |
| 5 |
Hb_001046_040 |
0.1166861128 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_044624_010 |
0.1176570461 |
- |
- |
- |
| 7 |
Hb_000363_250 |
0.117922385 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 8 |
Hb_001951_230 |
0.1200268105 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 9 |
Hb_001013_050 |
0.1200555979 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_002375_080 |
0.1210663946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004324_220 |
0.1264453459 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 12 |
Hb_003562_040 |
0.1275507735 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 13 |
Hb_002809_120 |
0.1286925248 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 14 |
Hb_001269_530 |
0.1324732741 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 15 |
Hb_175670_010 |
0.1332601616 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_001269_240 |
0.1337415256 |
- |
- |
aquaporin NIP6.1 family protein [Populus trichocarpa] |
| 17 |
Hb_002335_010 |
0.1366047568 |
- |
- |
hypothetical protein JCGZ_23475 [Jatropha curcas] |
| 18 |
Hb_000813_090 |
0.1368571108 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 19 |
Hb_004907_070 |
0.1372439591 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 20 |
Hb_000589_330 |
0.1373763164 |
- |
- |
kinase, putative [Ricinus communis] |