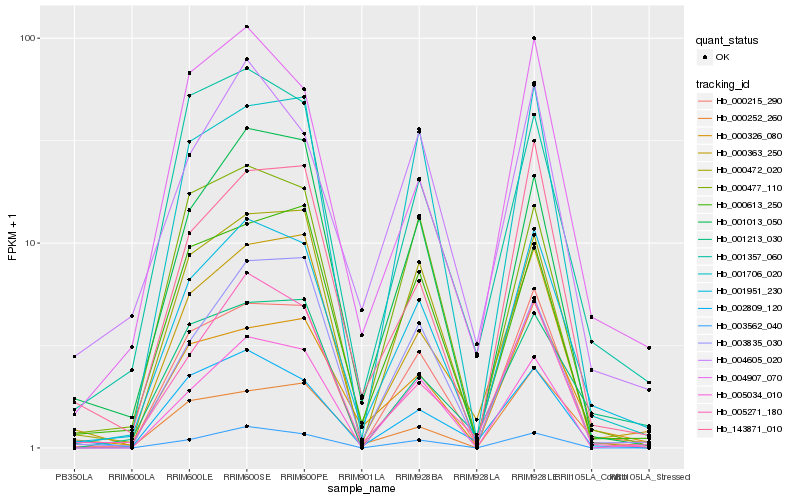
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_230 |
0.0 |
- |
- |
PREDICTED: phytochrome E isoform X2 [Jatropha curcas] |
| 2 |
Hb_000215_290 |
0.0978062059 |
transcription factor |
TF Family: NAC |
PREDICTED: protein BEARSKIN2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000326_080 |
0.1066594032 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_004907_070 |
0.1158304332 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 5 |
Hb_000363_250 |
0.1184660191 |
- |
- |
PREDICTED: pirin-like protein [Malus domestica] |
| 6 |
Hb_005271_180 |
0.1200268105 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 7 |
Hb_001357_060 |
0.1265805855 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 8 |
Hb_001013_050 |
0.1280669659 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_143871_010 |
0.1313568045 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 10 |
Hb_001213_030 |
0.1328962101 |
- |
- |
PAR1 protein [Theobroma cacao] |
| 11 |
Hb_000613_250 |
0.1344272796 |
- |
- |
PREDICTED: nudix hydrolase 2-like [Jatropha curcas] |
| 12 |
Hb_000472_020 |
0.1348787435 |
- |
- |
PREDICTED: uncharacterized protein LOC105641103 [Jatropha curcas] |
| 13 |
Hb_000252_260 |
0.135600637 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 14 |
Hb_001706_020 |
0.1358647375 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 15 |
Hb_003562_040 |
0.1363075301 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1-like [Jatropha curcas] |
| 16 |
Hb_003835_030 |
0.1365308683 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 17 |
Hb_002809_120 |
0.136986841 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 18 |
Hb_004605_020 |
0.1370176728 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 19 |
Hb_005034_010 |
0.1374982425 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_000477_110 |
0.1396739313 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein SLR1-like [Jatropha curcas] |