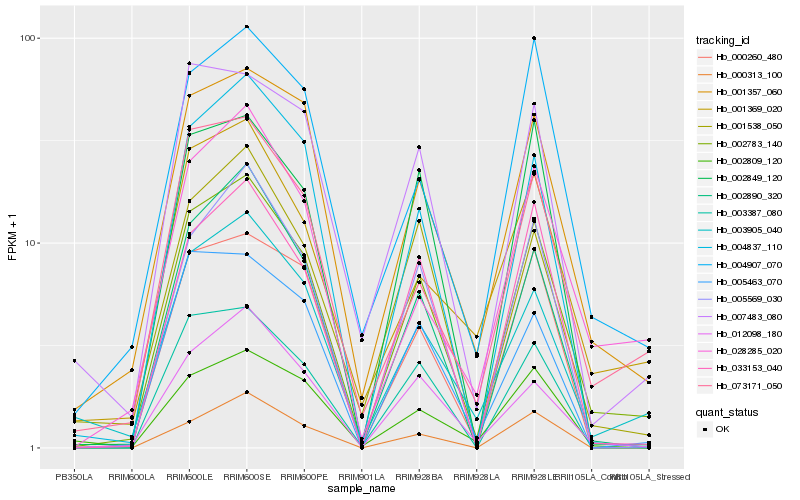
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 2 |
Hb_001357_060 |
0.1063955389 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 18-like [Cicer arietinum] |
| 3 |
Hb_002890_320 |
0.1143806601 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 4 |
Hb_033153_040 |
0.1146697538 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_005569_030 |
0.1150666912 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003387_080 |
0.1172908132 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_073171_050 |
0.1178334179 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 8 |
Hb_003905_040 |
0.1181730572 |
- |
- |
PREDICTED: heptahelical transmembrane protein 2 [Jatropha curcas] |
| 9 |
Hb_004837_110 |
0.1197319932 |
- |
- |
PREDICTED: MATE efflux family protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001538_050 |
0.1215634295 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 11 |
Hb_012098_180 |
0.1217925326 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 12 |
Hb_004907_070 |
0.1228781639 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 13 |
Hb_000260_480 |
0.1238641887 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |
| 14 |
Hb_002849_120 |
0.1287499586 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 15 |
Hb_028285_020 |
0.1302498565 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_002809_120 |
0.1312593094 |
- |
- |
Uncharacterized protein TCM_031281 [Theobroma cacao] |
| 17 |
Hb_007483_080 |
0.1313552785 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 18 |
Hb_001369_020 |
0.1326268725 |
- |
- |
PREDICTED: U-box domain-containing protein 35-like [Jatropha curcas] |
| 19 |
Hb_000313_100 |
0.1326486448 |
rubber biosynthesis |
Gene Name: CPT4 |
PREDICTED: rubber cis-polyprenyltransferase HRT2 [Jatropha curcas] |
| 20 |
Hb_005463_070 |
0.1338171726 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |