| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
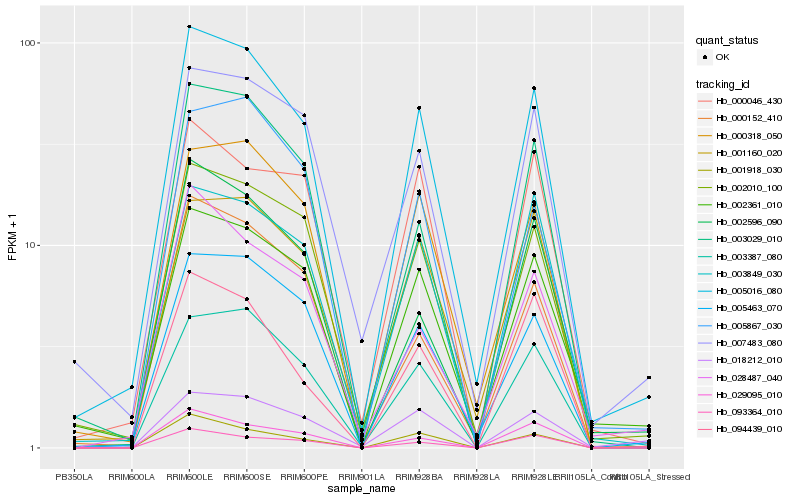
Hb_005016_080 |
0.0 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 2 |
Hb_002010_100 |
0.0782531477 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 3 |
Hb_002361_010 |
0.0810749672 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_005463_070 |
0.0873999398 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_000318_050 |
0.0920964841 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003387_080 |
0.0967791218 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001160_020 |
0.1068921862 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 8 |
Hb_007483_080 |
0.1071610653 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 9 |
Hb_003849_030 |
0.1107175696 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 10 |
Hb_001918_030 |
0.1115256719 |
- |
- |
- |
| 11 |
Hb_003029_010 |
0.1138135177 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002596_090 |
0.1257051313 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_093364_010 |
0.1258789222 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 14 |
Hb_000152_410 |
0.1262025069 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 15 |
Hb_000046_430 |
0.1266499526 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 16 |
Hb_018212_010 |
0.1288124769 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 17 |
Hb_029095_010 |
0.1295990268 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 18 |
Hb_028487_040 |
0.129986884 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 19 |
Hb_005867_030 |
0.133109389 |
- |
- |
PREDICTED: uncharacterized protein At4g00950 [Jatropha curcas] |
| 20 |
Hb_094439_010 |
0.133238536 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |