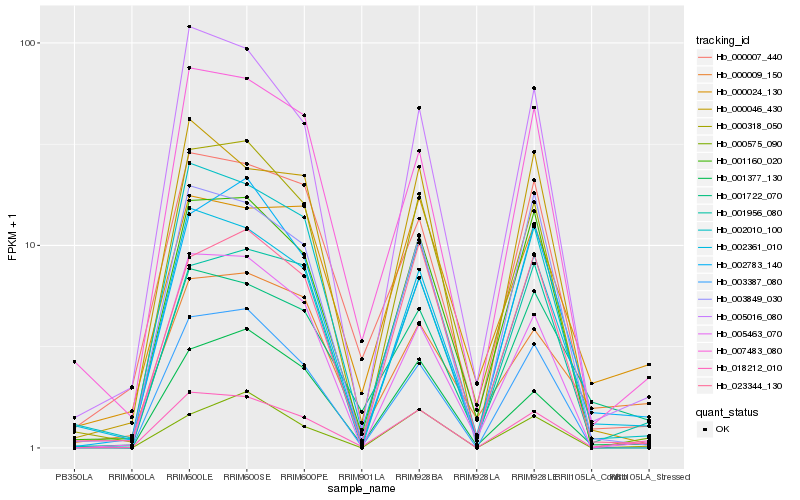
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018212_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 2 |
Hb_002361_010 |
0.1110855859 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_007483_080 |
0.1264944571 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 4 |
Hb_002010_100 |
0.1276645636 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 5 |
Hb_005016_080 |
0.1288124769 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 6 |
Hb_001956_080 |
0.1312039123 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 7 |
Hb_001722_070 |
0.1338988715 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 8 |
Hb_001377_130 |
0.1404653462 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 1-like [Jatropha curcas] |
| 9 |
Hb_003849_030 |
0.1431938122 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 10 |
Hb_000318_050 |
0.1444281559 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000046_430 |
0.1455174698 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 12 |
Hb_001160_020 |
0.1470962462 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 13 |
Hb_003387_080 |
0.1471771941 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000007_440 |
0.1484268885 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 15 |
Hb_000575_090 |
0.1491769148 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Populus euphratica] |
| 16 |
Hb_005463_070 |
0.1502336951 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_023344_130 |
0.1524305 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_002783_140 |
0.1528452123 |
- |
- |
PREDICTED: uncharacterized protein LOC105635332 [Jatropha curcas] |
| 19 |
Hb_000024_130 |
0.1545824064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000009_150 |
0.1579438 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |