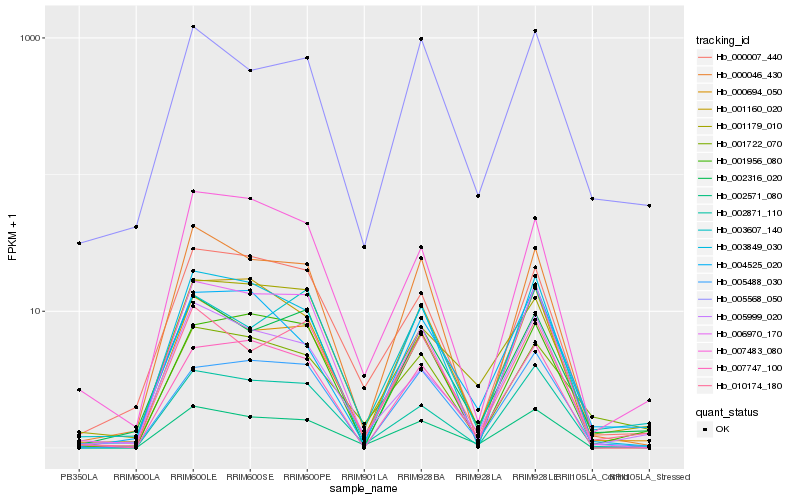
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002571_080 |
0.0 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |
| 2 |
Hb_005999_020 |
0.1163425587 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 3 |
Hb_000694_050 |
0.1197388167 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 4 |
Hb_010174_180 |
0.1201129263 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_002871_110 |
0.1266696106 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 6 |
Hb_001160_020 |
0.1309297488 |
transcription factor |
TF Family: NF-YA |
Nuclear transcription factor Y subunit A-3, putative [Ricinus communis] |
| 7 |
Hb_000007_440 |
0.1332765308 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 8 |
Hb_000046_430 |
0.1355746704 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 9 |
Hb_003849_030 |
0.1376049296 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 10 |
Hb_001179_010 |
0.1381417484 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 11 |
Hb_007747_100 |
0.1382323809 |
- |
- |
PREDICTED: probable F-box protein At1g60180 [Jatropha curcas] |
| 12 |
Hb_006970_170 |
0.1388031924 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_002316_020 |
0.1403091511 |
- |
- |
PREDICTED: uncharacterized protein LOC105640407 [Jatropha curcas] |
| 14 |
Hb_005488_030 |
0.1404479052 |
- |
- |
F3F9.11 [Arabidopsis lyrata subsp. lyrata] |
| 15 |
Hb_007483_080 |
0.1411967785 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 16 |
Hb_001722_070 |
0.1413310714 |
transcription factor |
TF Family: C2H2 |
PREDICTED: UBX domain-containing protein 6 [Jatropha curcas] |
| 17 |
Hb_001956_080 |
0.1417658423 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 14 [Jatropha curcas] |
| 18 |
Hb_005568_050 |
0.1488166348 |
- |
- |
ascorbate peroxidase [Hevea brasiliensis] |
| 19 |
Hb_003607_140 |
0.1496899282 |
- |
- |
3-ketoacyl-CoA thiolase B, putative [Ricinus communis] |
| 20 |
Hb_004525_020 |
0.1497788938 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 6-like [Jatropha curcas] |