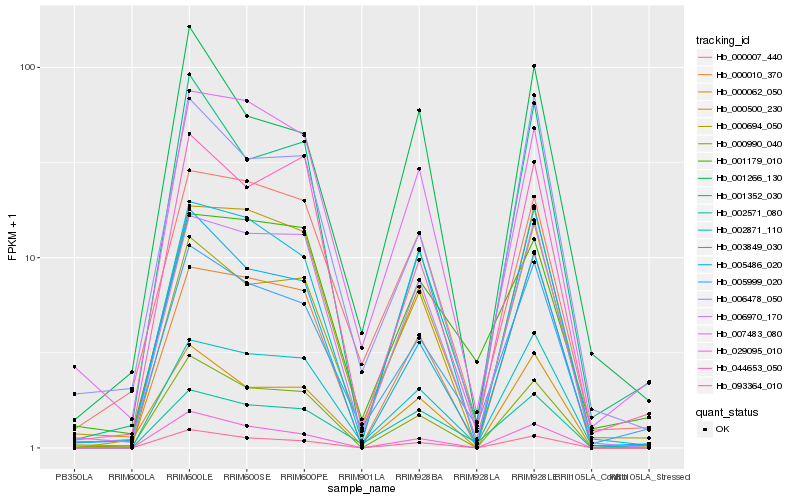
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005999_020 |
0.0 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 2 |
Hb_006478_050 |
0.1098937076 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007483_080 |
0.1135424197 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 4 |
Hb_002871_110 |
0.1136273731 |
transcription factor |
TF Family: ARF |
hypothetical protein POPTR_0014s12980g [Populus trichocarpa] |
| 5 |
Hb_002571_080 |
0.1163425587 |
- |
- |
secondary cell wall-related glycosyltransferase family 14 [Populus tremula x Populus tremuloides] |
| 6 |
Hb_005486_020 |
0.1165114816 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 7 |
Hb_000007_440 |
0.1194326276 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 8 |
Hb_006970_170 |
0.1212009934 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000500_230 |
0.1214947805 |
- |
- |
hypothetical protein RCOM_0634810 [Ricinus communis] |
| 10 |
Hb_000694_050 |
0.1224759816 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 11 |
Hb_000990_040 |
0.1262169898 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 12 |
Hb_044653_050 |
0.1275569644 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000010_370 |
0.1296795293 |
- |
- |
PREDICTED: uncharacterized protein LOC105640263 [Jatropha curcas] |
| 14 |
Hb_000062_050 |
0.1306563961 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_003849_030 |
0.1314673616 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 16 |
Hb_093364_010 |
0.1329319629 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 17 |
Hb_001352_030 |
0.1347814312 |
- |
- |
PREDICTED: uncharacterized protein LOC105636310 [Jatropha curcas] |
| 18 |
Hb_001179_010 |
0.1361088222 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 19 |
Hb_029095_010 |
0.1365477087 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 20 |
Hb_001266_130 |
0.1370060048 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |