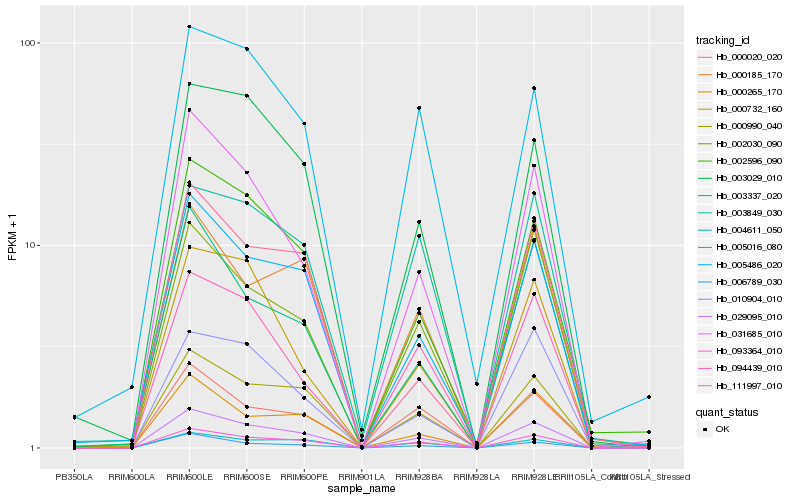
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029095_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 2 |
Hb_093364_010 |
0.0319397958 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 3 |
Hb_005486_020 |
0.0792917623 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 4 |
Hb_000990_040 |
0.0797021975 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 7 [Jatropha curcas] |
| 5 |
Hb_002596_090 |
0.0925571281 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_031685_010 |
0.0936273789 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_004611_050 |
0.0940996301 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 8 |
Hb_000020_020 |
0.1012031772 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 9 |
Hb_006789_030 |
0.1079362772 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 10 |
Hb_003029_010 |
0.1086109456 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002030_090 |
0.1086410815 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 12 |
Hb_094439_010 |
0.1141671424 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 13 |
Hb_000185_170 |
0.1175590343 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000265_170 |
0.1179824061 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like isoform X1 [Populus euphratica] |
| 15 |
Hb_010904_010 |
0.1207613879 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003337_020 |
0.1224298967 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 17 |
Hb_111997_010 |
0.1266924641 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 18 |
Hb_000732_160 |
0.1266957422 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 19 |
Hb_003849_030 |
0.1291701295 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 20 |
Hb_005016_080 |
0.1295990268 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |