| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
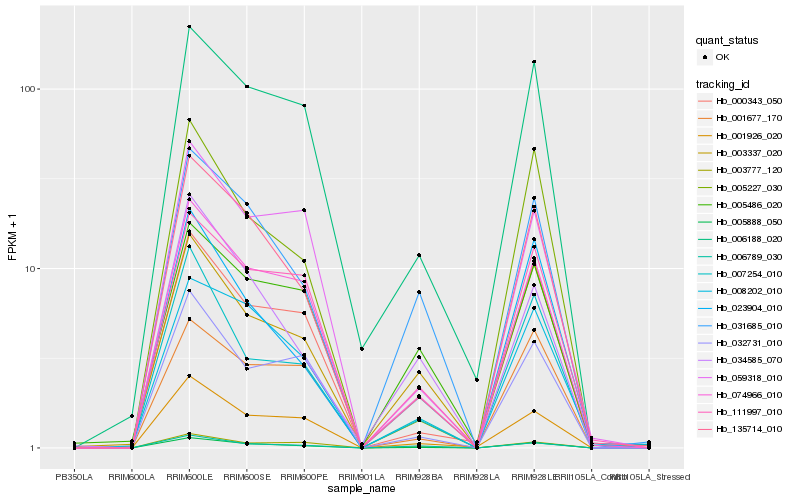
Hb_005888_050 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_135714_010 |
0.0612456657 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 3 |
Hb_074966_010 |
0.0761864056 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 4 |
Hb_001926_020 |
0.0941981337 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 5 |
Hb_008202_010 |
0.0967124364 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_031685_010 |
0.0985107789 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_006188_020 |
0.1014240959 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 8 |
Hb_111997_010 |
0.1045237573 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 9 |
Hb_003337_020 |
0.1058864626 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 10 |
Hb_000343_050 |
0.1088019972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003777_120 |
0.1159235995 |
- |
- |
- |
| 12 |
Hb_006789_030 |
0.1161017979 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 13 |
Hb_032731_010 |
0.1164779162 |
- |
- |
RING-H2 finger protein ATL5M precursor, putative [Ricinus communis] |
| 14 |
Hb_007254_010 |
0.1201893383 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 15 |
Hb_005227_030 |
0.1249123498 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 16 |
Hb_059318_010 |
0.1256111782 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 17 |
Hb_023904_010 |
0.127831549 |
- |
- |
- |
| 18 |
Hb_001677_170 |
0.1303753472 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 19 |
Hb_005486_020 |
0.1313502532 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 20 |
Hb_034585_070 |
0.1330676551 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |