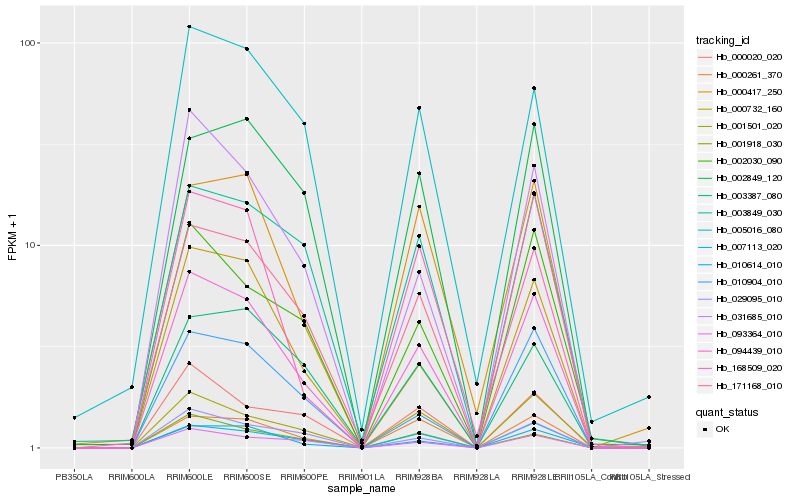
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_094439_010 |
0.0 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 2 |
Hb_000732_160 |
0.097704039 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 3 |
Hb_007113_020 |
0.1060477705 |
- |
- |
- |
| 4 |
Hb_010614_010 |
0.1080999431 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 5 |
Hb_031685_010 |
0.1100653255 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_029095_010 |
0.1141671424 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 7 |
Hb_093364_010 |
0.1165840624 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 8 |
Hb_002030_090 |
0.1208256343 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 9 |
Hb_010904_010 |
0.125704305 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 10 |
Hb_171168_010 |
0.1273396281 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001501_020 |
0.1285136842 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 12 |
Hb_003849_030 |
0.1315875204 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 13 |
Hb_000261_370 |
0.1318595721 |
- |
- |
- |
| 14 |
Hb_000020_020 |
0.132862571 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 15 |
Hb_005016_080 |
0.133238536 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 16 |
Hb_001918_030 |
0.1342905176 |
- |
- |
- |
| 17 |
Hb_000417_250 |
0.1367130939 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 18 |
Hb_002849_120 |
0.1386776326 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 19 |
Hb_003387_080 |
0.1443202844 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_168509_020 |
0.148409955 |
- |
- |
macrophage migration inhibitory factor family protein [Populus trichocarpa] |