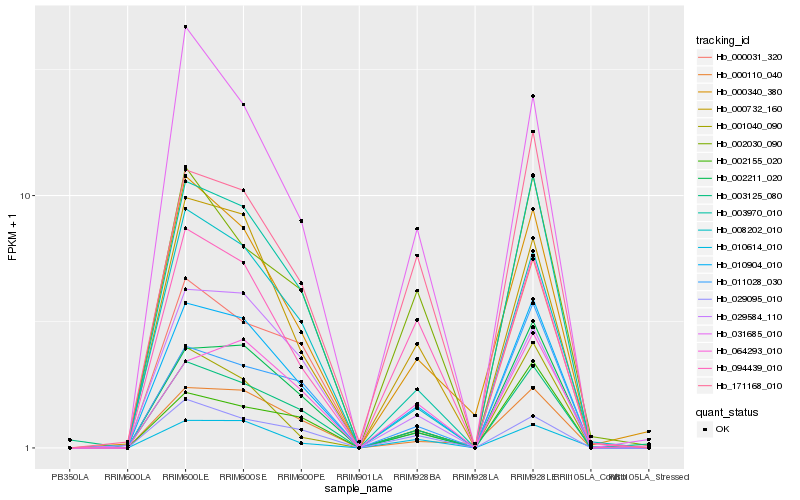
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010904_010 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 2 |
Hb_003970_010 |
0.0700851799 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_002211_020 |
0.0844129863 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 4 |
Hb_000732_160 |
0.1026805473 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_171168_010 |
0.1136417502 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 6 |
Hb_008202_010 |
0.1182169904 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000110_040 |
0.1189601124 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 8 |
Hb_029095_010 |
0.1207613879 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 9 |
Hb_000340_380 |
0.1256819392 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 10 |
Hb_094439_010 |
0.125704305 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 11 |
Hb_002030_090 |
0.1261971648 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 12 |
Hb_064293_010 |
0.1291644037 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_031685_010 |
0.1301927087 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_010614_010 |
0.1314482331 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 15 |
Hb_003125_080 |
0.1314691782 |
- |
- |
PREDICTED: F-box protein At5g50450 [Jatropha curcas] |
| 16 |
Hb_002155_020 |
0.1325595396 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 17 |
Hb_029584_110 |
0.1329377249 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 18 |
Hb_011028_030 |
0.1333002455 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000031_320 |
0.1336859104 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001040_090 |
0.1369346844 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |