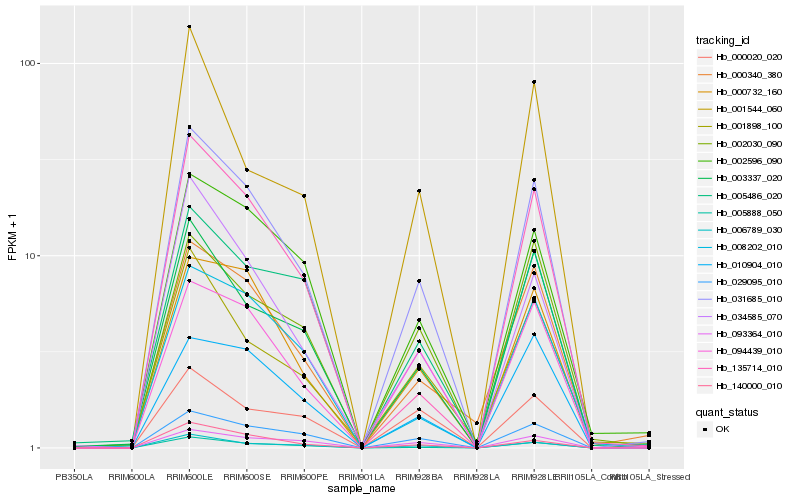
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031685_010 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_006789_030 |
0.093487373 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 3 |
Hb_029095_010 |
0.0936273789 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 4 |
Hb_000732_160 |
0.0970292687 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_005888_050 |
0.0985107789 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_003337_020 |
0.1074987414 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 7 |
Hb_094439_010 |
0.1100653255 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 8 |
Hb_140000_010 |
0.110966826 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 9 |
Hb_034585_070 |
0.1125918526 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 10 |
Hb_000340_380 |
0.1127069984 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 11 |
Hb_002596_090 |
0.1132830651 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_093364_010 |
0.1177225594 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 13 |
Hb_135714_010 |
0.1241650389 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 14 |
Hb_001898_100 |
0.1244721983 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 15 |
Hb_008202_010 |
0.126817233 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005486_020 |
0.1286864517 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 17 |
Hb_010904_010 |
0.1301927087 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002030_090 |
0.1337899181 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 19 |
Hb_001544_060 |
0.146703196 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000020_020 |
0.1470581021 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |