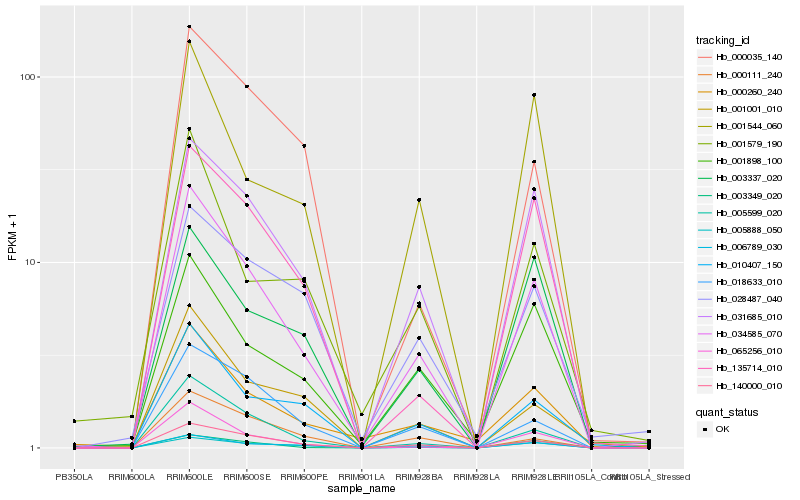
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034585_070 |
0.0 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 2 |
Hb_140000_010 |
0.0745753061 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_005599_020 |
0.1000813121 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 4 |
Hb_006789_030 |
0.1086589728 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 5 |
Hb_031685_010 |
0.1125918526 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_065256_010 |
0.1297868808 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 7 |
Hb_005888_050 |
0.1330676551 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_018633_010 |
0.1336978944 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_010407_150 |
0.133882422 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 10 |
Hb_000260_240 |
0.1486421054 |
- |
- |
hypothetical protein JCGZ_00622 [Jatropha curcas] |
| 11 |
Hb_135714_010 |
0.1488617137 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 12 |
Hb_001001_010 |
0.1493361534 |
- |
- |
PREDICTED: patatin-like protein 2 [Jatropha curcas] |
| 13 |
Hb_001898_100 |
0.1497714045 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 14 |
Hb_003349_020 |
0.1532690205 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_000111_240 |
0.1543817846 |
- |
- |
PREDICTED: glutamate receptor 2.8-like [Elaeis guineensis] |
| 16 |
Hb_001544_060 |
0.1627216895 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000035_140 |
0.1655478439 |
transcription factor |
TF Family: WRKY |
putative WRKY transcription factor family protein [Populus trichocarpa] |
| 18 |
Hb_003337_020 |
0.1694910504 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 19 |
Hb_001579_190 |
0.1701425384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_028487_040 |
0.1708707958 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |