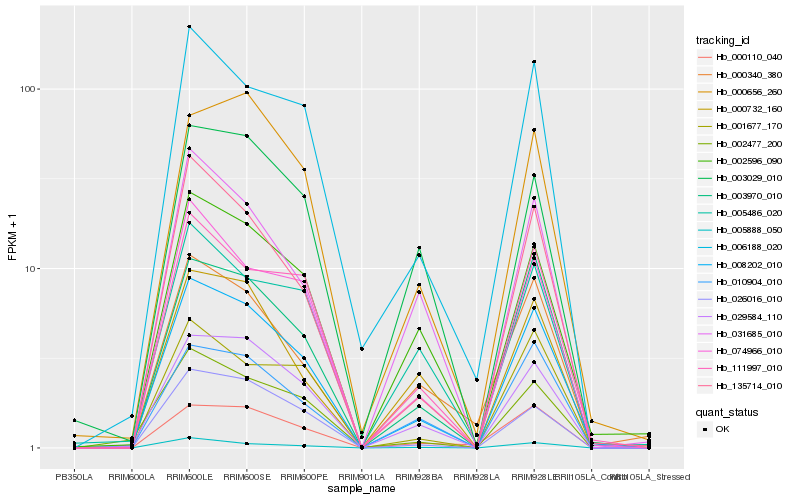
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008202_010 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_029584_110 |
0.0907060291 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 3 |
Hb_135714_010 |
0.0946909111 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 4 |
Hb_005888_050 |
0.0967124364 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_003970_010 |
0.098151859 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 6 |
Hb_002596_090 |
0.1026602922 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_111997_010 |
0.1026697788 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 8 |
Hb_006188_020 |
0.1084854679 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 9 |
Hb_074966_010 |
0.1123830536 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 10 |
Hb_026016_010 |
0.1166384649 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_010904_010 |
0.1182169904 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002477_200 |
0.1203063553 |
- |
- |
PREDICTED: uncharacterized protein LOC105631678 [Jatropha curcas] |
| 13 |
Hb_031685_010 |
0.126817233 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000110_040 |
0.1269528272 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 15 |
Hb_000656_260 |
0.1279921895 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 16 |
Hb_000340_380 |
0.129104722 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 17 |
Hb_000732_160 |
0.1297640356 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 18 |
Hb_001677_170 |
0.132992941 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 19 |
Hb_005486_020 |
0.1332200829 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 20 |
Hb_003029_010 |
0.1341656356 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |