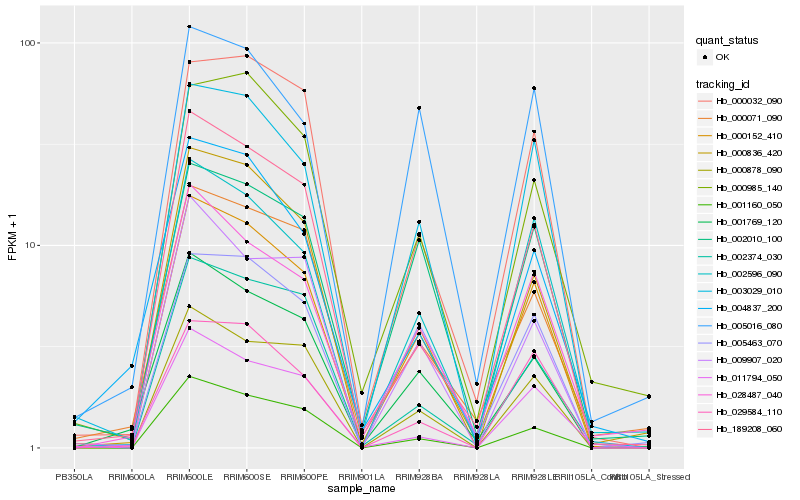
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_410 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 2 |
Hb_003029_010 |
0.0727126361 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002596_090 |
0.0925596997 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000878_090 |
0.0951082789 |
- |
- |
sucrose-phosphatase family protein [Populus trichocarpa] |
| 5 |
Hb_001769_120 |
0.0975803229 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 7A [Jatropha curcas] |
| 6 |
Hb_000836_420 |
0.0986555938 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_028487_040 |
0.0987940637 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 8 |
Hb_001160_050 |
0.1001305161 |
- |
- |
PREDICTED: long-chain-alcohol oxidase FAO1 [Jatropha curcas] |
| 9 |
Hb_189208_060 |
0.1007188788 |
- |
- |
PREDICTED: two-pore potassium channel 5 [Jatropha curcas] |
| 10 |
Hb_000032_090 |
0.1088627869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005463_070 |
0.1129063165 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_000071_090 |
0.1150350883 |
- |
- |
PREDICTED: probable protein phosphatase 2C 4 [Jatropha curcas] |
| 13 |
Hb_002010_100 |
0.1164925152 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 14 |
Hb_000985_140 |
0.1173106028 |
- |
- |
PREDICTED: uncharacterized protein LOC105643381 isoform X3 [Jatropha curcas] |
| 15 |
Hb_009907_020 |
0.117581417 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 16 |
Hb_002374_030 |
0.1177377487 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 isoform X1 [Jatropha curcas] |
| 17 |
Hb_029584_110 |
0.1190447135 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 18 |
Hb_011794_050 |
0.1214702372 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 19 |
Hb_005016_080 |
0.1262025069 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 20 |
Hb_004837_200 |
0.1292337422 |
- |
- |
hypothetical protein POPTR_0001s26450g [Populus trichocarpa] |