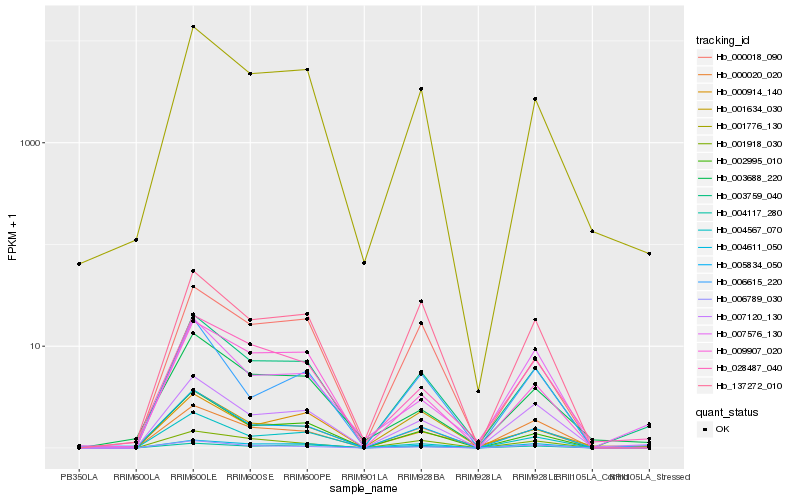
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003759_040 |
0.0 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 2 |
Hb_007120_130 |
0.0958038222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001776_130 |
0.111459223 |
- |
- |
metallothionein [Hevea brasiliensis] |
| 4 |
Hb_005834_050 |
0.1122271795 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 5 |
Hb_028487_040 |
0.1227624631 |
- |
- |
hypothetical protein POPTR_0010s11910g [Populus trichocarpa] |
| 6 |
Hb_002995_010 |
0.1235684603 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 7 |
Hb_000914_140 |
0.1248046283 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_003688_220 |
0.141887156 |
- |
- |
unknown [Populus trichocarpa] |
| 9 |
Hb_137272_010 |
0.1436180716 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 10 |
Hb_000018_090 |
0.1437444111 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_006789_030 |
0.1458349028 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 12 |
Hb_004611_050 |
0.1462004646 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 13 |
Hb_006615_220 |
0.1493389242 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_007576_130 |
0.150379767 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 15 |
Hb_004117_280 |
0.1506269031 |
- |
- |
hypothetical protein JCGZ_00361 [Jatropha curcas] |
| 16 |
Hb_001918_030 |
0.1510958397 |
- |
- |
- |
| 17 |
Hb_000020_020 |
0.1524900652 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 18 |
Hb_009907_020 |
0.1536715635 |
- |
- |
hypothetical protein CICLE_v10009323mg [Citrus clementina] |
| 19 |
Hb_004567_070 |
0.1543089208 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001634_030 |
0.1543699535 |
- |
- |
Receptor like protein 1, putative [Theobroma cacao] |