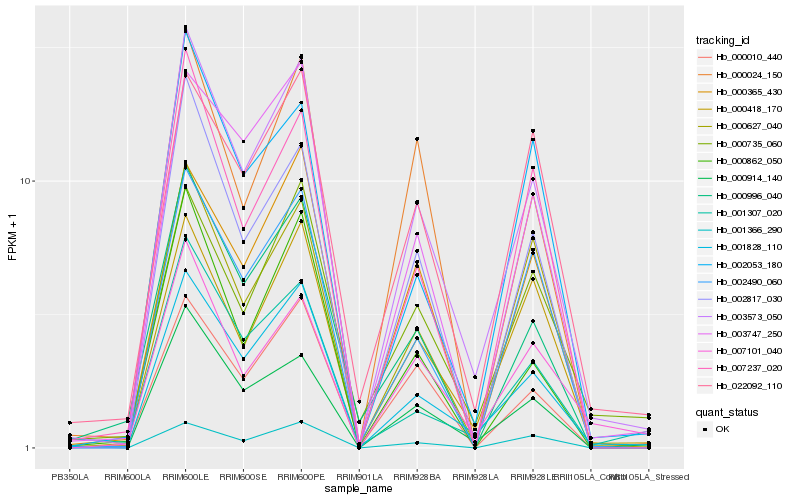
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003573_050 |
0.0 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 2 |
Hb_001828_110 |
0.0505101568 |
- |
- |
PREDICTED: lysine histidine transporter 1-like [Tarenaya hassleriana] |
| 3 |
Hb_000914_140 |
0.0896607458 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_002817_030 |
0.1091315697 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 5 |
Hb_000418_170 |
0.1115462332 |
- |
- |
PREDICTED: ABC transporter C family member 12-like isoform X4 [Populus euphratica] |
| 6 |
Hb_003747_250 |
0.1170808559 |
- |
- |
PREDICTED: potassium transporter 7 isoform X2 [Elaeis guineensis] |
| 7 |
Hb_002490_060 |
0.1195052263 |
- |
- |
PREDICTED: uncharacterized protein LOC105638743 [Jatropha curcas] |
| 8 |
Hb_000627_040 |
0.1207879146 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 9 |
Hb_000010_440 |
0.125154152 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 10 |
Hb_000365_430 |
0.1253278174 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g08570-like [Jatropha curcas] |
| 11 |
Hb_002053_180 |
0.1273287327 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 12 |
Hb_007237_020 |
0.1274566036 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 13 |
Hb_000862_050 |
0.1284581004 |
transcription factor |
TF Family: GRAS |
DELLA protein RGL1, putative [Ricinus communis] |
| 14 |
Hb_000024_150 |
0.128772012 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 15 |
Hb_007101_040 |
0.1292521947 |
- |
- |
PREDICTED: MLP-like protein 423 [Populus euphratica] |
| 16 |
Hb_001366_290 |
0.1297790421 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000735_060 |
0.12995036 |
- |
- |
PREDICTED: U-box domain-containing protein 9 [Jatropha curcas] |
| 18 |
Hb_001307_020 |
0.1306904199 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 19 |
Hb_000996_040 |
0.1311314618 |
- |
- |
hypothetical protein POPTR_0003s12500g [Populus trichocarpa] |
| 20 |
Hb_022092_110 |
0.1337351296 |
- |
- |
PREDICTED: probable protein phosphatase 2C 52 [Jatropha curcas] |