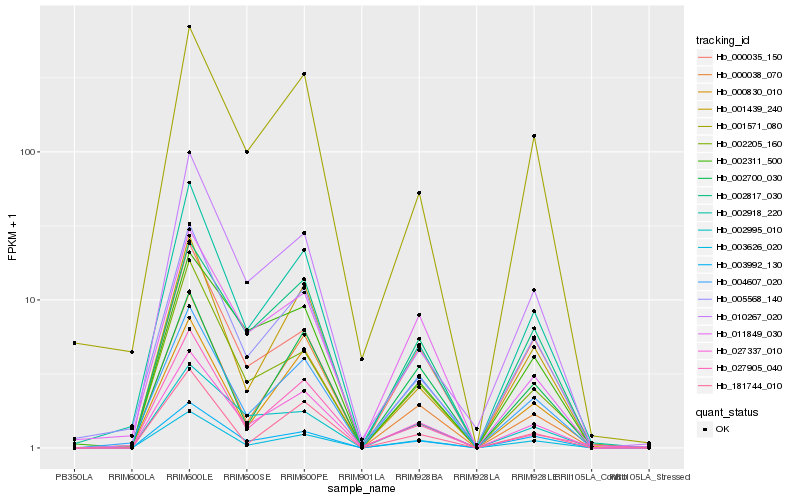
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027337_010 |
0.0 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 2 |
Hb_003992_130 |
0.0901146003 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_002817_030 |
0.0938122917 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |
| 4 |
Hb_003626_020 |
0.0944121053 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At5g24080 [Jatropha curcas] |
| 5 |
Hb_002918_220 |
0.0944130895 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 6 |
Hb_001571_080 |
0.0993423934 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 7 |
Hb_005568_140 |
0.1081793779 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_004607_020 |
0.1108480342 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-like [Jatropha curcas] |
| 9 |
Hb_001439_240 |
0.1128109666 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 10 |
Hb_002311_500 |
0.1172168236 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 11 |
Hb_002995_010 |
0.1194320475 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 12 |
Hb_000035_150 |
0.1204827981 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 13 |
Hb_011849_030 |
0.1204961945 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002205_160 |
0.1222543847 |
- |
- |
chalcone isomerase [Hibiscus cannabinus] |
| 15 |
Hb_000038_070 |
0.1224136123 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000830_010 |
0.1244339748 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_181744_010 |
0.1262138065 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 18 |
Hb_027905_040 |
0.1275496315 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: E2F transcription factor-like E2FF [Jatropha curcas] |
| 19 |
Hb_010267_020 |
0.1295623504 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 20 |
Hb_002700_030 |
0.1309055739 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |