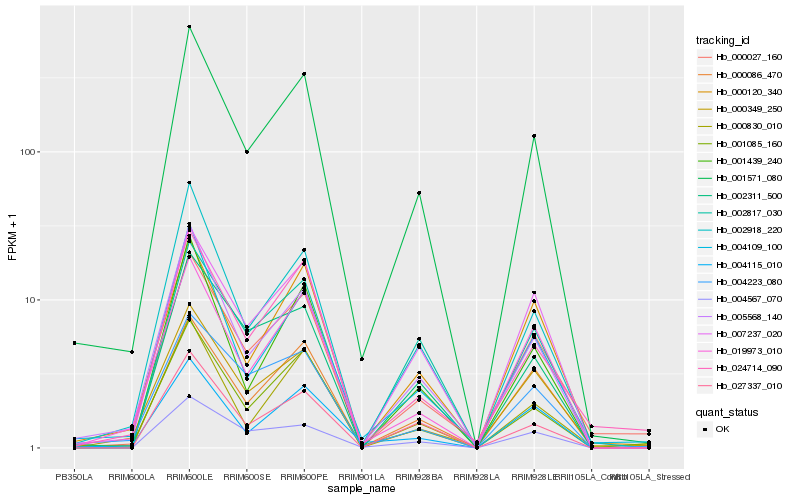
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001571_080 |
0.0 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 2 |
Hb_005568_140 |
0.0730484635 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_001085_160 |
0.0817108185 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 4 |
Hb_002918_220 |
0.0865008387 |
- |
- |
PREDICTED: uncharacterized protein LOC105138896 [Populus euphratica] |
| 5 |
Hb_007237_020 |
0.0961189594 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 6 |
Hb_027337_010 |
0.0993423934 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 7 |
Hb_000027_160 |
0.1022502193 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 8 |
Hb_019973_010 |
0.1048252 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000830_010 |
0.1057847176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002311_500 |
0.1063401389 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 11 |
Hb_004109_100 |
0.1066028807 |
- |
- |
Interactor of constitutive active rops 1 isoform 1 [Theobroma cacao] |
| 12 |
Hb_000086_470 |
0.1075528139 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 13 |
Hb_000349_250 |
0.1087403884 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 14 |
Hb_001439_240 |
0.1096604129 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0017s12230g [Populus trichocarpa] |
| 15 |
Hb_004223_080 |
0.1118479859 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_024714_090 |
0.1146704647 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 17 |
Hb_000120_340 |
0.1149746758 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 18 |
Hb_004115_010 |
0.1152157058 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HDG11 [Jatropha curcas] |
| 19 |
Hb_004567_070 |
0.1158261839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002817_030 |
0.1158740602 |
- |
- |
hypothetical protein POPTR_0001s39195g [Populus trichocarpa] |