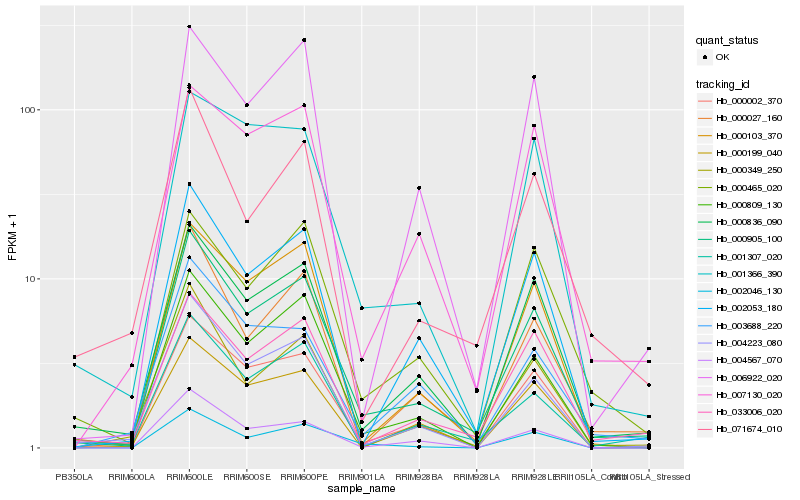
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000905_100 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000836_090 |
0.0948911863 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 3 |
Hb_000809_130 |
0.1064961573 |
- |
- |
PREDICTED: peroxidase 10 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000002_370 |
0.1067420787 |
- |
- |
PREDICTED: uncharacterized protein LOC105632103 [Jatropha curcas] |
| 5 |
Hb_002053_180 |
0.1084681334 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 6 |
Hb_000027_160 |
0.1106400387 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 7 |
Hb_033006_020 |
0.1148482568 |
- |
- |
Aspartate aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_001307_020 |
0.1175405388 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 9 |
Hb_000103_370 |
0.1185928323 |
- |
- |
PREDICTED: protein argonaute 10 [Jatropha curcas] |
| 10 |
Hb_004223_080 |
0.1195545268 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_002046_130 |
0.1234938409 |
- |
- |
PREDICTED: UDP-sugar transporter sqv-7-like [Jatropha curcas] |
| 12 |
Hb_000349_250 |
0.1263426885 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 13 |
Hb_004567_070 |
0.1302443735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007130_020 |
0.1349656743 |
- |
- |
- |
| 15 |
Hb_000465_020 |
0.1366621618 |
- |
- |
PREDICTED: uncharacterized protein LOC105632280 [Jatropha curcas] |
| 16 |
Hb_006922_020 |
0.1384542656 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 34 [Jatropha curcas] |
| 17 |
Hb_071674_010 |
0.1393798623 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 18 |
Hb_000199_040 |
0.1411746775 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001366_390 |
0.1415281637 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 20 |
Hb_003688_220 |
0.1418916289 |
- |
- |
unknown [Populus trichocarpa] |