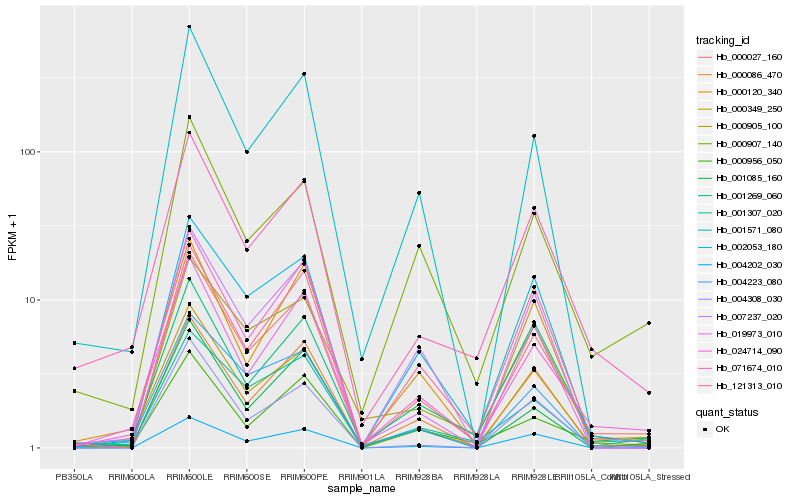
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000027_160 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000349_250 |
0.075684372 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g12460 [Jatropha curcas] |
| 3 |
Hb_024714_090 |
0.0898758176 |
rubber biosynthesis |
Gene Name: SRPP3 |
PREDICTED: stress-related protein [Jatropha curcas] |
| 4 |
Hb_002053_180 |
0.0976027584 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_001571_080 |
0.1022502193 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At5g64080 [Jatropha curcas] |
| 6 |
Hb_000086_470 |
0.1072018638 |
- |
- |
hypothetical protein JCGZ_17645 [Jatropha curcas] |
| 7 |
Hb_000905_100 |
0.1106400387 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH130-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001307_020 |
0.112589345 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 9 |
Hb_007237_020 |
0.1147073499 |
- |
- |
PREDICTED: protein trichome birefringence-like 23 [Jatropha curcas] |
| 10 |
Hb_001085_160 |
0.1157297816 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 11 |
Hb_071674_010 |
0.1159128472 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 12 |
Hb_000120_340 |
0.1168131977 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 13 |
Hb_019973_010 |
0.1200074734 |
- |
- |
PREDICTED: probable methyltransferase PMT18 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001269_060 |
0.120933674 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g80640 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004223_080 |
0.1211480741 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_004202_030 |
0.1212277622 |
- |
- |
PREDICTED: uncharacterized protein LOC103449702 [Malus domestica] |
| 17 |
Hb_000907_140 |
0.1220490939 |
- |
- |
PREDICTED: annexin D5-like [Jatropha curcas] |
| 18 |
Hb_004308_030 |
0.1278541791 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 19 |
Hb_121313_010 |
0.1279201146 |
- |
- |
PREDICTED: endoglucanase 24-like [Jatropha curcas] |
| 20 |
Hb_000956_050 |
0.1285963602 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |